In this issue of Blood, Yan et al study the anatomy of azanuceoside methylation reversal.1
The azanucleoside analogues 5-azacytidine and decitabine improve hematopoiesis in approximately half of treated myelodysplastic syndrome patients2,3 ; in high-risk patients, 5-azactytidine improves overall survival compared with other typical clinical practice.2 Because both drugs are active inhibitors of DNA methyltransferase and can reactivate tumor suppressor genes silenced through cytosine methylation in CpG-rich promoter regions (so-called CpG islands), conventional wisdom assumes that these drugs exert their clinical effects through similar mechanisms. In fact, these drugs are commonly referred to as hypomethylating agents. Despite the biologically attractive dogma, attempts to identify specific genes or groups of genes whose methylation reversal can be associated with or predictive of clinical response have not resulted in clear causal relationships.4-6
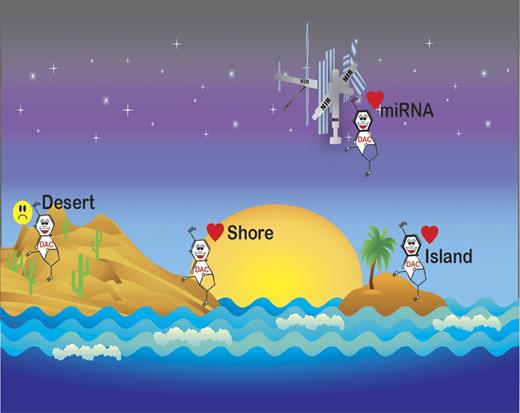
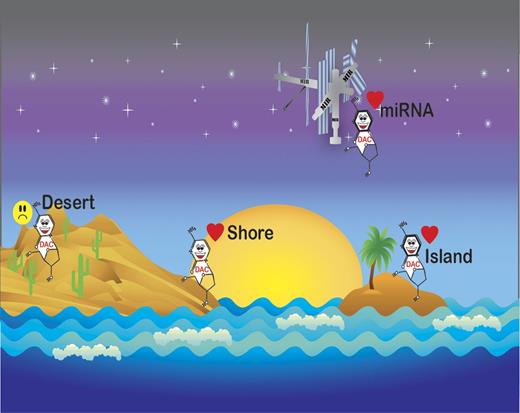
Recently, the epigenome has been recognized as increasingly complex.7 While it has long been known that cancer genomes are hypomethylated outside of CpG islands, the significance of epigenetic changes in noncoding regions constitutes an area of intensive research due to the availability of newer sequencing technology. Yan and colleagues combined capture of methylated DNA with next generation sequencing (MethylCap-seq) to examine bone marrow samples obtained from patients during their first cycle of treatment with decitabine.1 Methylation on day 25 of treatment was compared with pretreatment bone marrow. Hypomethylation at 25 days after treatment was significant in genomic regions associated with CpG islands, CpG island shores, CpG inlands, miRNA-associated CpG islands and promoters, RefSeq genes, and RefSeq gene–associated CpG islands (see figure). Clinical responders and nonresponders had similar changes in methylation in each region; however, nonresponders demonstrated a smaller extent of methylation reversal. Differentially methylated regions significantly clustered on the ends of all but 5 chromosomes.
Decitabine-induced methylation reversal occurs in CpG Islands, shores, and mIR-associated promoters, but not gene deserts.
Decitabine-induced methylation reversal occurs in CpG Islands, shores, and mIR-associated promoters, but not gene deserts.
This study should be seen as a demonstration of feasibility. Only 16 patients were included, and the only true discriminator between patients who subsequently developed clinical responses and those who did not was the extent of methylation reversal1 ; decrease in global methylation can be measured using much simpler and inexpensive techniques! Nonetheless, as the significance of methylation changes in different genomic regions becomes clearer in both normal and malignant cells, exploration of differential methylation changes in specific regions in response to azanucleosides may lead to a better understanding of the mechanisms underlying the clinical activity of these drugs. Ultimately, such information may help in 2 directions: (1) clinically, understanding mechanism may lead to design of better drugs. And (2), at a more basic level, understanding the significance of perturbing methylation patterns of specific regions of the genome may better our appreciation of epigenomic organization and cellular regulation.
Conflict-of-interest disclosure: The authors declare no competing financial interests. ■