Abstract
MicroRNAs (miRs) are involved in many aspects of normal and malignant hematopoiesis, including hematopoietic stem cell (HSC) self-renewal, proliferation, and terminal differentiation. However, a role for miRs in the generation of the earliest stages of lineage committed progenitors from HSCs has not been identified. Using Dicer inactivation, we show that the miR complex is not only essential for HSC maintenance but is specifically required for their erythroid programming and subsequent generation of committed erythroid progenitors. In bipotent pre-MegEs, loss of Dicer up-regulated transcription factors preferentially expressed in megakaryocyte progenitors (Gata2 and Zfpm1) and decreased expression of the erythroid-specific Klf1 transcription factor. These results show a specific requirement for Dicer in acquisition of erythroid lineage programming and potential in HSCs and their subsequent erythroid lineage differentiation, and in particular indicate a role for the miR complex in achieving proper balance of lineage-specific transcriptional regulators necessary for HSC multilineage potential to be maintained.
Introduction
MicroRNAs (miRs) are an abundant class of endogenous small noncoding RNAs that regulate gene expression, such as gene silencing and degradation, uncovering a new layer of regulatory control of gene expression in many organisms.1 To date, almost 1000 different miRs have been identified, playing important roles in proliferation, differentiation, and apoptosis.2
miR biogenesis requires successive processing events carried out by the RNAses Drosha and Dicer. Thus, in the absence of Dicer, essentially all miRs are depleted, making Dicer deletion a method by which their role in biological processes may be assessed. Recently, it was suggested that Dicer is involved in regulation of the hematopoietic stem cell (HSC) niche3 as well as in intrinsic regulation of HSCs themselves.4 Individual miRs have been implicated in differentiation of mature blood cell lineages, such as B cells,5 T cells,6 and granulocytes.7 Recently, miR-141/4518 and miR-1919 were implicated in terminal erythroid differentiation, and in vitro studies have suggested that miR-150 is involved in the development of megakaryocytes from megakaryocyte/erythroid progenitors.10 However, a requirement for Dicer in regulating the early stages of myelo/erythroid lineage commitment remains to be demonstrated.
By deleting Dicer from the hematopoietic system, we show that Dicer is not only crucial for maintenance of HSCs but is selectively required for generation of the most primitive erythroid committed progenitors, whereas megakaryocyte and myeloid lineage commitment remains intact.
Methods
Mice
Gene expression analysis
Microarray analysis was performed as previously described.14 Microarray data have been deposited in the ArrayExpress database (www.ebi.ac.uk/arrayexpress; accession no. E-MTAB-674). Quantitative PCR analysis was performed using the BioMark Dynamic Array platform (Fluidigm) and TaqMan Gene Expression Assays (Applied Biosystems; supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
In vitro progenitor assays
Evaluation of erythroid (E) and myeloid (GM) potential was performed as described,15 and megakaryocyte (Mk) potential with the Megacult assay (StemCell Technologies) according to the manufacturer's instructions (supplemental Methods).
FACS
FACS analysis of peripheral blood and BM stem and progenitor cells was done on a LSRII analyzer and sorts on a FACSAriaII (BD Biosciences). For further details, see supplemental Methods.
Statistical analysis
Statistical significance was calculated using Mann-Whitney t test.
Results and discussion
Loss of erythroid transcriptional priming in Dicer-deficient HSCs
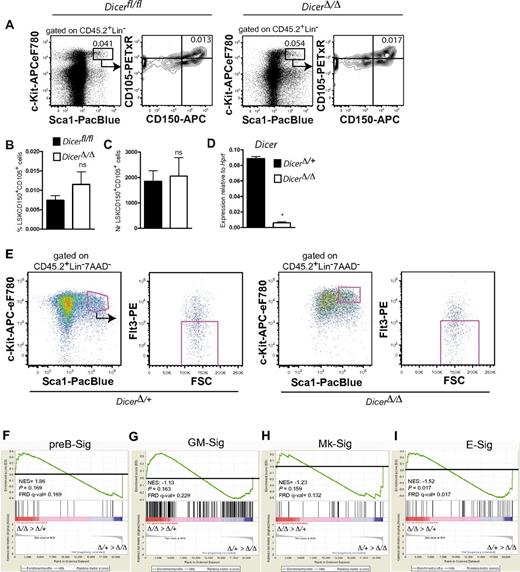
Previous reports suggested that miRs control HSC numbers.4 Through competitive transplantation experiments, using 2 different Cre lines, we found Dicer to be essential for maintenance of HSCs (supplemental Figure 1). To assess whether loss of Dicer from HSCs affects their lineage transcriptional priming we noncompetitively transplanted Dicerfl/fl;Mx1Cre and control Dicerfl/fl BM cells. BM cellularity was reduced > 2-fold in DicerΔ/Δ mice 13 days after deletion (supplemental Figure 2A). However, at this time point, phenotypic LSKCD105+CD150+ HSCs were present at normal levels (Figure 1A-C), even though Dicer mRNA was efficiently deleted (Figure 1D). Using global gene profiling of DicerΔ/+ and DicerΔ/Δ LSKFlt3− HSCs (Figure 1E) and gene set enrichment analysis,16 we observed that while lymphoid (pre-B), myeloid (GM), and megakaryocyte (Mk) lineage programming was not significantly affected by Dicer deletion (Figure 1F-H), there was a marked overall down-regulation of erythroid (E) genes in DicerΔ/Δ HSCs (Figure 1I). Importantly, because Dicer-deleted LSKCD105+CD150+ HSCs were present at normal levels at this time point (Figure 1A-B), any observed changes in transcriptional lineage priming in HSCs, or output of distinct progenitors, are unlikely in this setting to be because of depletion of HSCs.
Dicer regulates erythroid lineage transcriptional priming in HSCs. Lethally irradiated (9 Gy) wild-type CD45.1 mice were transplanted noncompetitively with 6 million BM cells from control (Dicerfl/fl or Dicerfl/+;Mx1Cre) or DicerΔ/Δ (Dicerfl/fl;Mx1Cre) mice. At least 4 weeks after transplantation, Cre-mediated recombination was induced with polyriboinosinic acid/polyribocytidylic acid (polyI:C), by intraperitoneal injections of 200 μg polyI:C every second day, 3 injections in total. (A) Representative FACS profiles of HSCs (LSKCD105+CD150+) in Dicerfl/fl and DicerΔ/Δ mice, respectively. Numbers in FACS profiles indicate percentage within indicated gate, relative to total BM cells. (B-C) Mean (± SEM) frequency relative to total BM cells (B; 10 or 11 mice/group, analyzed individually) and absolute number per 2 tibiae (C) of LSKCD105+CD150+ HSCs in Dicerfl/fl and DicerΔ/Δ mice. n = 4 or 5 mice/group analyzed individually. (D) Deletion efficiency of Dicer from 100-200 sorted LSKCD150+ HSCs. n = 6 mice/group. *P < .05. (E) Sorting strategy for isolation of CD45.2+LSKFlt3− cells from DicerΔ/+ and DicerΔ/Δ mice. FACS profiles are from kit-enriched samples. (F-I) Gene set enrichment analysis comparison of pre-B cell (preB-Sig; F), GM (GM-Sig; G), megakaryocyte (Mk-Sig; H), and erythroid (E-Sig; I) associated gene expression between DicerΔ/+ and DicerΔ/Δ LSKFlt3− HSCs. NES indicates normalized enrichment score; FDR, false discovery rate q value; and P, nominal P value. Two biological replicates were analyzed/genotyped in 2 separate experiments, using Affymetrix Mouse Genome 430 Version 2.0 arrays.
Dicer regulates erythroid lineage transcriptional priming in HSCs. Lethally irradiated (9 Gy) wild-type CD45.1 mice were transplanted noncompetitively with 6 million BM cells from control (Dicerfl/fl or Dicerfl/+;Mx1Cre) or DicerΔ/Δ (Dicerfl/fl;Mx1Cre) mice. At least 4 weeks after transplantation, Cre-mediated recombination was induced with polyriboinosinic acid/polyribocytidylic acid (polyI:C), by intraperitoneal injections of 200 μg polyI:C every second day, 3 injections in total. (A) Representative FACS profiles of HSCs (LSKCD105+CD150+) in Dicerfl/fl and DicerΔ/Δ mice, respectively. Numbers in FACS profiles indicate percentage within indicated gate, relative to total BM cells. (B-C) Mean (± SEM) frequency relative to total BM cells (B; 10 or 11 mice/group, analyzed individually) and absolute number per 2 tibiae (C) of LSKCD105+CD150+ HSCs in Dicerfl/fl and DicerΔ/Δ mice. n = 4 or 5 mice/group analyzed individually. (D) Deletion efficiency of Dicer from 100-200 sorted LSKCD150+ HSCs. n = 6 mice/group. *P < .05. (E) Sorting strategy for isolation of CD45.2+LSKFlt3− cells from DicerΔ/+ and DicerΔ/Δ mice. FACS profiles are from kit-enriched samples. (F-I) Gene set enrichment analysis comparison of pre-B cell (preB-Sig; F), GM (GM-Sig; G), megakaryocyte (Mk-Sig; H), and erythroid (E-Sig; I) associated gene expression between DicerΔ/+ and DicerΔ/Δ LSKFlt3− HSCs. NES indicates normalized enrichment score; FDR, false discovery rate q value; and P, nominal P value. Two biological replicates were analyzed/genotyped in 2 separate experiments, using Affymetrix Mouse Genome 430 Version 2.0 arrays.
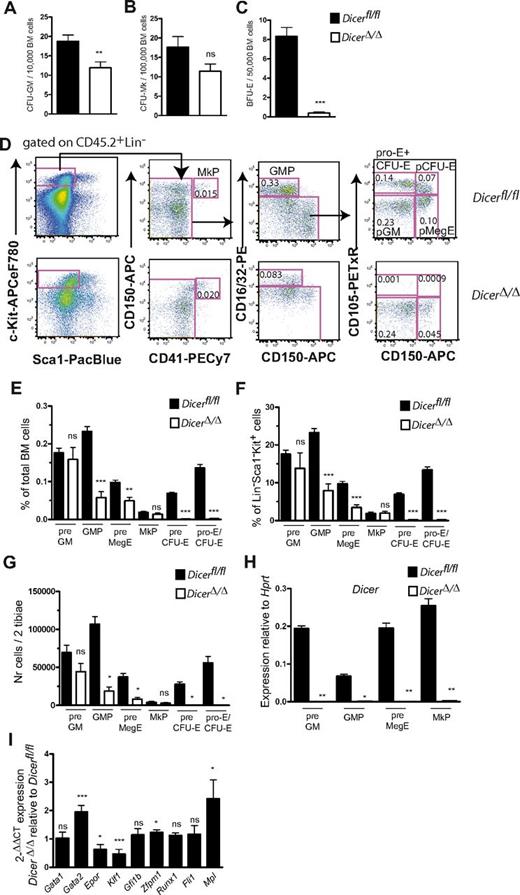
In vitro colony assays revealed normal Mk colony numbers and mildly reduced CFU-GM, in striking contrast to the complete loss of BFU-Es in DicerΔ/Δ BM, both upon direct deletion (Figure 2A-C) and deletion after noncompetitive transplantation (supplemental Figure 2B-E). To establish at which stage of the erythroid differentiation pathway Dicer is required, detailed FACS staging of myelo/erythroid progenitors in DicerΔ/Δ BM was performed.18 This analysis demonstrated a specific and complete loss of the earliest erythroid committed progenitors (pre-CFU-Es, CFU-Es) and a reduction in the preceding pre-MegE progenitors in DicerΔ/Δ BM (Figure 2D-G), whereas MkPs and pre-GMs were present at normal levels, indicating unimpaired megakaryocytic and myeloid lineage commitment. Consistent with the lower number of CFU-GM, granulocyte-monocyte progenitor (GMPs) were reduced (Figure 2D-G). Specific deletion of Dicer in myeloid progenitors, using Cebpa-Cre, does not result in impaired GMP formation.19 The observed reduction of GMPs may therefore reflect that earlier, and possibly more efficient, Dicer depletion is able to reveal a role for miRs in myeloid lineage progression.
Dicer is critical for the earliest stages of the erythroid progenitor compartment. (A-C) Mean (± SEM) CFU-GM (A), CFU-Mk (B), and BFU-E (C) colonies at 8 to 9 days of culture from unfractionated BM cells isolated 12 days after intraperitoneal injections of 200 μg polyI:C every second day, 3 injections in total, of 11-day-old Dicerfl/fl (n = 4) and Dicer fl/fl;Mx1Cre mice (n = 8). Each mouse was analyzed in quadruplicate. (D) Representative FACS profiles of myelo/erythroid progenitor cells from lethally irradiated CD45.1 wild-type mice transplanted noncompetitively with Dicerfl/fl and DicerΔ/Δ (CD45.2) BM cells, analyzed 12 days after polyI:C treatment. Numbers indicate percentage within indicated gate, of total BM cells. (E-F) Mean (± SEM) frequencies of test cell-derived myelo/erythroid progenitors in transplanted mice. n = 10 or 11 mice/group, analyzed individually. (G) Mean (± SEM) numbers of test cell-derived myelo/erythroid progenitors in transplanted mice. n = 4 or 5 mice/group, analyzed individually. (H) Dicer deletion efficiency in pre-GMs, GMPs, pre-MegEs, and MkPs isolated from mice transplanted with DicerΔ/Δ BM cells and injected with polyI:C 12 days prior to analysis. Data are mean (± SEM) values from cells purified from 2 different pools of mice/genotype, 2 or 3 replicate wells/pool. (I) Megakaryocyte/erythroid gene expression in 100-200 pre-MegEs isolated from Dicerfl/fl and DicerΔ/Δ mice. Mean (± SEM) expression in DicerΔ/Δ relative to control Dicerfl/fl pre-MegEs, calculated using the 2−ΔΔCT method.17 Data generated from sorts performed on 6 cohorts of mice/genotype (2 or 3 mice/cohort), 13 or 14 technical replicates, 3 experiments in total. *P < .05. **P < .01. ***P < .001. ns indicates not significant. Statistics refer to comparison between DicerΔ/Δ mice and Dicerfl/fl control mice.
Dicer is critical for the earliest stages of the erythroid progenitor compartment. (A-C) Mean (± SEM) CFU-GM (A), CFU-Mk (B), and BFU-E (C) colonies at 8 to 9 days of culture from unfractionated BM cells isolated 12 days after intraperitoneal injections of 200 μg polyI:C every second day, 3 injections in total, of 11-day-old Dicerfl/fl (n = 4) and Dicer fl/fl;Mx1Cre mice (n = 8). Each mouse was analyzed in quadruplicate. (D) Representative FACS profiles of myelo/erythroid progenitor cells from lethally irradiated CD45.1 wild-type mice transplanted noncompetitively with Dicerfl/fl and DicerΔ/Δ (CD45.2) BM cells, analyzed 12 days after polyI:C treatment. Numbers indicate percentage within indicated gate, of total BM cells. (E-F) Mean (± SEM) frequencies of test cell-derived myelo/erythroid progenitors in transplanted mice. n = 10 or 11 mice/group, analyzed individually. (G) Mean (± SEM) numbers of test cell-derived myelo/erythroid progenitors in transplanted mice. n = 4 or 5 mice/group, analyzed individually. (H) Dicer deletion efficiency in pre-GMs, GMPs, pre-MegEs, and MkPs isolated from mice transplanted with DicerΔ/Δ BM cells and injected with polyI:C 12 days prior to analysis. Data are mean (± SEM) values from cells purified from 2 different pools of mice/genotype, 2 or 3 replicate wells/pool. (I) Megakaryocyte/erythroid gene expression in 100-200 pre-MegEs isolated from Dicerfl/fl and DicerΔ/Δ mice. Mean (± SEM) expression in DicerΔ/Δ relative to control Dicerfl/fl pre-MegEs, calculated using the 2−ΔΔCT method.17 Data generated from sorts performed on 6 cohorts of mice/genotype (2 or 3 mice/cohort), 13 or 14 technical replicates, 3 experiments in total. *P < .05. **P < .01. ***P < .001. ns indicates not significant. Statistics refer to comparison between DicerΔ/Δ mice and Dicerfl/fl control mice.
Blood analysis 12 days after deletion revealed a significant reduction in red blood cells, hemoglobin, and hematocrit, but not platelet numbers (supplemental Figure 2F), consistent with Dicer being required for erythroid, but not megakaryocyte lineage commitment. White blood cells were also reduced in Dicer-deficient mice (supplemental Figure 2F), most probably reflecting the reported role of Dicer in maturation of granulocytes and lymphocytes.7-9
Importantly, analysis of myelo-erythroid progenitors in DicerΔ/Δ BM confirmed the absence of Dicer mRNA (Figure 2H). Consistent with this, the Dicer-dependent miR-148a, but not the Dicer-independent miR-451, was depleted in both HSCs and pre-MegEs (supplemental Figure 2G). Expression of common Mk/E transcription factors Gata1, Gfi1b, Runx1, and Fli1 was unaffected in DicerΔ/Δ pre-MegEs (Figure 2I), whereas expression of the Mk-related Gata2, Zfpm1, and Mpl was increased, and the E genes Epor and Klf1 decreased in pre-MegEs. Notably, at the normal Mk/E bifurcation, Gata2 and Zfpm1 are up-regulated in MkPs, whereas Klf1 is up-regulated in pre-CFU-Es.18 Likewise, Mpl, the key cytokine receptor for early Mk development, was up-regulated in DicerΔ/Δ pre-MegEs. These data provide support for miRs acting as developmental switches during cell fate determination by fine-tuning the lineage-specific transcriptomes. The changes in lineage-specific transcription factor and cytokine receptor expression are probably the result of a wide-ranging deregulation of miR-controlled genes. Using gene set enrichment analysis, we observed that 203 of 219 gene sets defined by specific miR seeds being present in the 3′-untranslated region showed up-regulation of gene expression in DicerΔ/Δ HSCs, with 122 of 219 of these being significantly up-regulated (P < .05; false discovery rate < 0.25; no gene sets significantly down-regulated; supplemental Table 1). These data confirm a general role for miRs in suppression of mRNA expression but also indicate that a large number of miRs may contribute to the observed phenotype, although it needs to be considered that pre-miR accumulation may also play a role. Furthermore, recently it was suggested that Dicer may also regulate gene expression by binding chromatin directly, suggesting that Dicer might also affect hematopoiesis through siRNA-independent mechanisms.20
So far, miRs have not been implicated in the early stages of erythroid progenitors, but rather in maturation of erythroid cells, such as miR-144/4518 and miR-191.9 Furthermore, miR-150 has been suggested to drive the differentiation of megakaryocyte/erythroid progenitors into the Mk lineage at the expense of the erythroid lineage.10 The results obtained here show that Dicer-deficient hematopoiesis already at the stem cell level is depleted of erythroid gene expression; and in line with this, DicerΔ/Δ HSCs subsequently fail to generate the earliest stages of erythroid lineage-committed progenitors, whereas myeloid and megakaryocytic lineage commitment seems unaffected. This both identifies the miR complex as selectively required for the generation of the earliest stages of erythroid commitment and supports the proposed role of HSC lineage priming in subsequent commitment events.21-24
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Biomedical Services at Oxford University and Theresa Furey and Helen Ferry for technical assistance, Paul Collier and Vladimir Benes (EMBL, Heidelberg) for help with miR quantitative RT-PCR analysis, and Elizabeth Zuo (Stanford University) for help with affymetrix analysis.
This work was supported by the United Kingdom MRC (Strategic Awards and Program Grants, S.E.W.J. and C.N.) and the Swedish Research Council (HematoLinne Strategic Award). A.J.M. was supported by the Leukemia and Lymphoma Research Senior Bennett Fellowship. D.A. was supported by the Leukemia and Lymphoma Research Foundation.
Authorship
Contribution: S.E.W.J., C.N., V.B.C., and N.B.-V. designed and conceptualized the overall research, analyzed the data, and wrote the manuscript; V.B.C., N.B.-V., P.S.W., M.L., A.J.M., L.M., S.L., T.B.-J., and D.A. performed experiments; V.B.C., S.M., and C.N. performed Affymetrix analysis; and D.O. provided Dicer conditional knockout mice.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Claus Nerlov, Institute for Stem Cell Research, University of Edinburgh, Edinburgh EH9 16UU, United Kingdom; e-mail: cnerlov@staffmail.ed.ac.uk; and Sten Eirik W. Jacobsen, Weatherall Institute of Molecular Medicine, University of Oxford, Oxford OX3 9DS, United Kingdom; e-mail: sten.jacobsen@imm.ox.ac.uk.
References
Author notes
N.B.-V. and V.B.C. contributed equally to this study.
S.E.W.J. and C.N. contributed equally to this study.