In this issue of Blood, Burkhart et al provide an impressive near complete quantitative analysis of the proteins in platelets, encompassing almost 4000 proteins and representing approximately 85% of the total protein content.1
Since the beginning of proteomics research on blood platelets, the aim has been to count all the proteins in these anucleate cells to better understand their role in health and disease and to improve antithrombotic medication. This is the biochemist's approach: one needs to know which molecules are expressed in a cell for optimally modulating its function. Especially in platelets with limited mRNA translation, knowledge of the protein content and profile might be more relevant than information on the mRNA transcriptome. The present paper from Burkhart et al is a major and decisive step forward to achieve this goal.1 However, there are still some challenges ahead.
During the past decade, several research groups have generated preliminary lists of the platelet proteome and of posttranslational modifications of platelet proteins, such as the sites of phosphorylation, palmitoylation, or N-glycosylation. Together, they identified more than 2000 proteins and more than 1000 posttranslational modifications in resting and activated platelets.2-4 Because these lists show no more than partial overlap with those of the platelet transcriptome, the field has been waiting for an updated, more extensive proteomic analysis. Interest in platelet proteomics is also boosted by indications that the platelet protein content varies with age and in patients with cardiovascular or other diseases.5-7 Even antiplatelet medication (clopidogrel, aspirin) may provoke changes in platelet protein expression levels. Hence, it is relevant not only to find the missing platelet proteins, but also to know the precise amounts of these proteins.
Burkhart and colleagues have managed to identify more than 4000 proteins in human platelets, and determined the copy numbers of approximately 3700 of those. The authors used well-purified, washed platelets, with no more than 1% of the proteins derived from plasma. The proteins quantified in this paper covered 80% to 85% of the entire platelet protein content. Intra-subject variation of the analysis was small, with 85% of the proteins showing no significant difference between consecutive blood donations. Remarkably, the inter-subject variance was also small for low as well as high copy numbers, with 85% of the quantified proteins showing (almost) no variation among healthy donors. Hence, despite the delicacy of the process of platelet shedding from megakaryocytes and the relatively fast turnover of circulating platelets, their proteome appears to be surprisingly stable. Another remarkable finding is that the protein profile does not correlate at all with earlier published transcriptome analyses, implicating that—at least in anucleate platelets—the protein composition is not a mere reflection of the mRNA content. Clearly, the quantitative protein list has many applications. It allows calculating stoichiometries between subunits of protein complexes and protein isoforms and, thus, will help to understand the biochemistry of these proteins. For instance, the finding that 3 phospholipase Cβ isoforms are present at similar copy numbers implies that all 3 will have relevant roles in agonist-induced Ca2+ signaling.8 The protein list contains many other surprises; for example, a relatively high expression of ubiquitination proteins in platelets.
Not disputing the major achievements made by Burkhart et al, there is still room for improvement. Proteomics analysis starts with trypsin digestion of a cell lysate, which gives specific problems. For instance, this procedure may result in underestimation of glycoproteins and proteins with high hydrophobicity or with multiple transmembrane domains. In addition, other proteins (eg, proteases) may form trypsin digestion fragments that due to their size or sequence are difficult to analyze, while protein isoforms may produce identical or indefinable sets of peptide fragments. This explains why in the new list G-protein–coupled receptors and other important membrane proteins are underrepresented. In addition, it is still problematic to quantify proteins with copy numbers below 500 per platelet. Specific enrichment procedures need to be employed in this case.
The procedure of protein quantification applied in the article by Burkhart et al uses a new method for conversion of mass spectrometry peaks to protein copy numbers. The authors use the so-called normalized spectral abundance factor (NSAF),1 which is a normalization procedure developed by the Washburn group, and corrects for the size of a given protein.9 Although the right control experiments were performed, further adaptation may be needed to precisely determine the amounts of proteins with difficult trypsin cleavage patterns. This may explain deviations between current expression numbers with those of other publications for several (membrane) proteins (eg, CD9), glycoprotein Ibβ, integrin α2, and gelsolin. Conceivably, the quantification can be improved by using fine-tuned normalization procedures or specific sets of calibration proteins. The next challenge now is to come to a fully calibrated quantitative protein database.
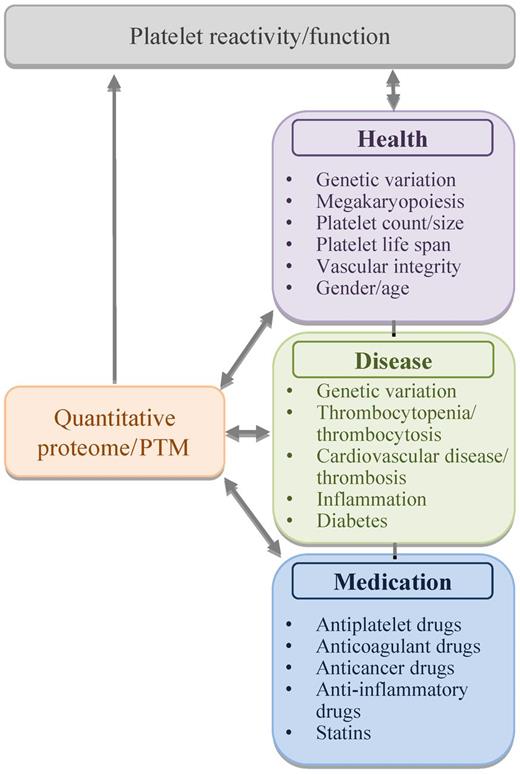
Yet, the current possibility to assess an almost complete platelet proteome with relatively small blood samples opens new avenues for research and diagnosis. The underlying concept is that protein expression levels and posttranslational modifications in platelets are controlled by multiple physiologic processes in health and disease, and are modulated by medication (see figure). Conversely, the composition and modification of these proteins could affect these physiologic processes. What the precise consequences are of variation in the platelet protein pattern will become clear in the near future.
The platelet protein mix as playmaker of platelet function. Protein expression levels and posttranslational modifications (PTM) may regulate and can be regulated by various physiologic processes in health and disease, via effects on platelet function and reactivity.
The platelet protein mix as playmaker of platelet function. Protein expression levels and posttranslational modifications (PTM) may regulate and can be regulated by various physiologic processes in health and disease, via effects on platelet function and reactivity.
Conflict-of-interest disclosure: The authors declare no competing financial interests. ■