Abstract
We analyzed the mutational hotspot region of SRSF2 (Pro95) in 275 cases with chronic myelomonocytic leukemia (CMML). In addition, ASXL1, CBL, EZH2, JAK2V617F, KRAS, NRAS, RUNX1, and TET2 mutations were investigated in subcohorts. Mutations in SRSF2 (SRSF2mut) were detected in 47% (129 of 275) of all cases. In detail, 120 cases had a missense mutation at Pro95, leading to a change to Pro95His, Pro95Leu, Pro95Arg, Pro95Ala, or Pro95Thr. In 9 cases, 3 new in/del mutations were observed: 7 cases with a 24-bp deletion, 1 case with a 3-bp duplication, and 1 case with a 24-bp duplication. In silico analyses predicted a damaging character for the protein structure of SRSF2 for all mutations. SRSF2mut was correlated with higher age, less pronounced anemia, and normal karyotype. SRSF2mut and EZH2mut were mutually exclusive, but SRSF2mut was associated with TET2mut. In the total cohort, no effect of SRSF2mut on survival was observed. However, in the RUNX1mut subcohort, SRSF2 Pro95His had a favorable effect on overall survival. This comprehensive mutation analysis found that 93% of all patients with CMML carried at least 1 somatic mutation in 9 recurrently mutated genes. In conclusion, these data show the importance of SRSF2mut as new diagnostic marker in CMML.
Introduction
Chronic myelomonocytic leukemia (CMML) is a clonal hematopoietic malignancy that can be characterized by features of both a myelodysplastic syndrome (MDS) and a myeloproliferative neoplasm (MPN). Therefore, the World Health Organization classification of 2008 assigned CMML to the mixed category MDS/MPN.1 A further characteristic feature is the wide heterogeneity of clinical presentations and course, leading to variable prognosis. Beside cytologic criteria for diagnosis, the only genetic criterion, until recently, was the absence of the BCR-ABL1 fusion transcript. The number of blasts in the peripheral blood (PB) and bone marrow (BM) is a prognostic factor dividing CMML cases into 2 morphologic categories: CMML-1 with fewer than 5% blasts in PB or 10% in BM, and CMML-2 with 5%-19% blasts in PB or 10%-19% in BM.1 Median overall survival (OS) is approximately 20 months in CMML-1 and 15 months in CMML-2, but wide variations exist.2 In approximately 15%-30% of patients with CMML, the disease evolves into acute myeloid leukemia (AML).1,2 On the basis of patient characteristics of 213 patients Onida et al defined a scoring system for CMML, named M.D. Anderson (MDA) prognostic score, stratifying patients with CMML in the 4 subgroups: low, intermediate-1, intermediate-2, and high risk. The level of risk is defined by 4 scores assigned by the following variables: hemoglobin levels below 12 g/dL, lymphocyte count higher than 2.5 × 109/L, presence of circulating immature myeloid cells, and bone marrow blasts 10% or more.3
Most patients show a normal karyotype in the CMML cells, and only 20%-40% show clonal cytogenetic abnormalities.1 Such and coworkers investigated 414 patients with CMML to evaluate the prognostic effect of cytogenetic abnormalities and identified 3 risk categories.4 A normal karyotype or loss of the Y-chromosome as a sole abnormality represent the low-risk group; trisomy 8, abnormalities of chromosome 7, or a complex karyotype (defined as 3 or more abnormalities) were related to the high-risk group. All other abnormalities were assigned to the intermediate-risk category.
In contrast to cytogenetic aberrations, several molecular gene mutations recently have been found to be frequent in CMML (resulting in overall mutation frequencies of > 55%5-8 ); but, unfortunately, none of these alterations is specific for CMML. Gene mutations identified in CMML cases affect different cellular targets and processes, such as RUNX19 (transcriptional regulation); isocitrate dehydrogenases IDH1/210 (metabolism); or KRAS, NRAS,11,12 CBL,13 and JAK214 (tyrosine-signaling pathways). TET2,15 DNMT3A,16 ASXL1,17 UTX,18 and EZH219 contribute in the broadest sense to epigenetic regulatory mechanisms. All of the cytogenetic changes and molecular mutations have been associated with the pathogenesis of CMML but do not fully explain leukemogenesis.
Thus far, mutations in several of these genes already show prognostic relevance. To date, EZH2 is the best molecularly analyzed gene in CMML and implies an unfavorable prognosis.5 Mutation of ASXL1 correlates with evolution to AML and a shorter OS.20 The effect of TET2 mutations remains controversial; in patients with MDS it is associated with a favorable outcome,1,21 and in CMML different studies found favorable to adverse clinical courses for it.6,22 Mutations in RUNX1 clearly correlate with a poor outcome in patients with MDS and patients with AML.23,24
Previously, we have investigated 81 CMML cases and analyzed the mutation frequency of a number of genes that were found to be recurrently mutated in CMML. These comprehensive studies resulted in an overall mutation frequency of 82%,5,6 indicating that there is a certain percentage of patients with unknown molecular alterations.
More recently, an additional cellular process was found to be altered in MDS. A whole-exome sequencing approach of 29 MDS specimens and their normal controls detected mutations in several components of the splicing machinery (ie, spliceosome; such as SF3B1 and U2AF1), mostly involved in 3′-splice site recognition. In this context a new candidate gene, SRSF2 (serine/arginine-rich splicing factor 2, also known as SC35, a classic member of the SR-protein family), was identified in close cooperation with our laboratory.7 Members of the SR-protein family function in constitutive and alternative splicing. They contain a RNA recognition motif (RRM) for binding to RNA and a arginine/serine-rich (RS) domain for interaction with other SR-proteins (Figure 1). As a component of the spliceosome, SRSF2 binds to exonic splicing enhancers, preventing exon skipping and ensuring the correct linear order of exons in spliced mRNA.25,26 In our recent study, mutations within the SRSF2 sequence occurred exclusively at position 95 (Pro95), located in a linker sequence between the 2 functional RRM and RS domains. SRSF2 was found to be most frequently mutated in CMML (28%), less frequently in MDS without increased ring sideroblasts (12%), and to some extent in refractory anemia with ring sideroblasts (6%) and AML/MDS (7%). It was rarely seen to be mutated in MPN (2%) or de novo AML (1%).7
To characterize further the genetic defects of CMML, we analyzed the frequency of SRSF2 mutations, their coincidence with other mutations, and their prognostic relevance in a large cohort of 275 cases.
Methods
Patient cohort
In total, 275 cases with CMML were analyzed. All cases were validated on peripheral blood and/or bone marrow smears according to standards of the World Health Organization1 and included in all cases May-Grünwald-Giemsa staining, as well as myeloperoxidase, nonspecific esterase, and iron stains.27 The cohort comprised 189 men and 86 women with a median age of 72.8 years (range, 21.9-93.3 years). Eighty-one patients who have been published previously by our group except for SRSF2 entered the cohort.5,6 There is no overlap with the CMML cohort analyzed in Yoshida et al.7 Cytogenetic analyses were performed after short-term culture. Karyotypes were analyzed after G-banding and were described according to the International System for Human Cytogenetic Nomenclature (1995 guidelines).28 Further parameters are given in Table 1. All patients gave their consent for genetic analyses and the use of laboratory results for research purposes. The study design adhered to the tenets of the Declaration of Helsinki and was approved by our institutional review board before its initiation.
Sequencing analyses
Isolation of mononuclear cells, DNA and mRNA extraction, and random primed cDNA synthesis were performed as described previously.29 A 187-bp fragment, containing the mutational hotspot region of SRSF2 around Pro95, was amplified with the GC-RICH PCR system (Roche Applied Science) from either genomic DNA (n = 201) or cDNA templates (n = 74), using the following primers: SRSF2-for, TTCGCCTTCGTTCGCTTT; SRSF2-rev, TCCGGCGTCCGTAGCCA. The single amplicon was analyzed by Sanger sequencing in all cases with the use of BigDye Term v1.1 cycle sequencing chemistry (Applied Biosystems). Estimation of the mutational load was based on the electropherograms of the forward and reverse reactions. In addition, in 10 cases the mutational load was confirmed by next-generation sequencing, showing the correlation of both methods (supplemental Figure 2B, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Additional mutational data obtained by Sanger sequencing, next-generation deep amplicon sequencing,30 or melting curve analyses were available in subcohorts and are described methodically elsewhere: ASXL1 exon 12 (n = 261),31 CBL (n = 274),6,32 EZH2 (n = 208),5 IDH1/2 (n = 82),5 JAK2V617F (n = 275),33 KRAS codons 12/13 and 61 (n = 266),6 NRAS codons 12/13 and 61 (n = 273),6,34 RUNX1 (n = 274),6 and TET2 (n = 160).6 The coding sequence of SF3B1 (n = 171) was analyzed by Sanger sequencing. U2AF1 Ser34 and Gln157 (n = 265) were analyzed by melting curve analyses.
In silico analyses
For protein structure prediction, we used the Robetta prediction server (http://robetta.bakerlab.org).35 In first iteration, we applied Robetta to predict models for the known RRM domain (2KN4.pdb) of the SRSF2 wild-type (wt) protein. On the basis of the resulting model, the 3-dimensional full model option was applied to obtain a complete model of SRSF2. Next, we repeated these steps to generate full models for our detected novel mutations. The altered protein sequences were submitted to Robetta, and resulting full models were compared with the SRSF2 wild-type model. For each submitted sequence, we selected the best model based on a manual validation process of the RRM domain. Finally, to analyze the differences between the best resulting models we calculated the Cα-Cα distances.36 For a more detailed report, see supplemental Methods.
Statistical analyses
Statistical analyses were performed with SPSS version 19.0.0 (SPSS Inc); the reported P values are 2-sided.
Survival curves were calculated for OS according to Kaplan-Meier and compared with the 2-sided log-rank test. OS was the time from diagnosis to death or last follow-up. Follow-up data were available in 180 cases, which were included in survival analyses. Results were considered significant at P < .05. Adjustment for multiple testing was not done. Dichotomous variables were compared between different groups with the use of the chi-square test and continuous variables by Student t test.
Results
Characterization of 275 patients with CMML
According to the classification of the World Health Organization, the 275 patients were categorized as 193 cases of CMML-1 and 82 cases of CMML-2. Morphologic features of monocytes and monoblasts and erythroid dysplastic changes are given in supplemental Methods. On the basis of biologic parameters 61 patients were categorized to the MDA score,3 with 10 patients belonging to the low-risk group, 15 to the intermediate-1 group, 25 to the intermediate-2 group, and 11 to the high-risk group.
Cytogenetic analyses were performed in 269 of 275 cases (in 6 cases, no metaphases were available). As typical in CMML a majority of patients had a normal karyotype (71%; 190 of 269), yet 29% (79 of 269) showed an aberrant karyotype. Within the aberrant karyotype group of 79 patients, a loss of the Y-chromosome (n = 13), chromosome 7 aberrations (n = 9), and a trisomy 8 (n = 26) were the most frequent abnormalities (for further parameters, see Table 1). Therefore, 203 cases belong to the low-risk category, whereas 27 belong to the intermediate- and 39 to the high-risk categories, defined by Such et al.4
Characterization and frequency of SRSF2 mutations
To analyze the mutation frequency of SRSF2 in our CMML cohort of 275 patients (Table 1), we investigated the sequence of an amplicon covering the mutation hotspot codon Pro95. Alterations of Pro95 or adjacent sequences were detected in 47% (129 of 275) of all cases. Mutation frequencies were similar in CMML-1 (47%; 91 of 193) and CMML-2 (46%; 38 of 82). In detail, 119 cases had a missense mutation leading to a change of Pro95 to 1 of the following 5 residues: p.Pro95His (n = 56), p.Pro95Leu (n = 38), p.Pro95Arg (n = 23), p.Pro95Ala (n = 1), and p.Pro95Thr (n = 1). In all cases, an estimated mutation load of 30%-50% in accordance with a heterozygous mutation status was detected. One additional case showed 2 different mutations p.[Arg94Pro;Pro95His] in a subset of 50% each. Next-generation sequencing validated it as a mono-allelic mutation.
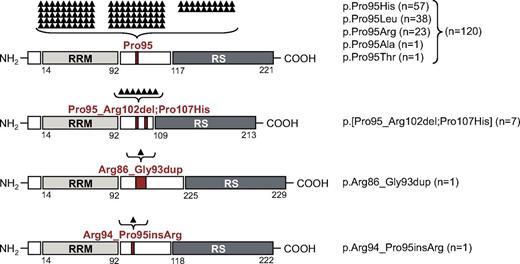
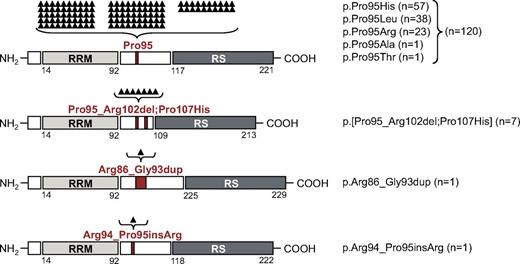
Interestingly, beyond the previously described missense mutations leading to alterations of Pro95,7 3 new in/del mutations were observed, affecting the immediate neighboring amino acids (AA) of Pro95. In 7 cases a deletion of 24 bp with a start in the codon of Pro95 resulted in deletion of 8 AAs, ranging from Pro95 to Arg102. All of these 7 cases showed an additional missense mutation at Pro107 (p.[Pro95_Arg102del;Pro107His]). Furthermore, 1 single case showed a 24-bp duplication of the AA Arg86 to Gly93 (p.Arg86_Gly93dup), and another sample had a 3-bp duplication that resulted in an insertion of arginine between Arg94 and Pro95 (p.Arg94_Pro95insArg). None of these mutations led to a frameshift. Buccal swab controls of 2 patients, carrying the p.[Pro95_Arg102del;Pro107His] mutation, were SRSF2 wild-type. Furthermore, 1 patient obtained this mutation during disease course. The National Institutes of Health dbSNP databases37 as well as the National Heart, Lung, and Blood Institute Exome variant server both report no missense single nucleotide polymorphisms for the analyzed region (AA 86-107), indicating that these novel mutations are somatic mutations and no germline polymorphisms. Figure 1 gives a schematic overview of the protein organization (based on information given by UniProtKB Q01130) and the mutation type, localization, and frequency. SRSF2-mutated sequences are shown in supplemental Figure 2A.
Schematic overview of SRSF2 protein organization, mutation type, and mutation frequency. SRSF2 consists of an RRM (AA 14-92; light gray box), a linker region (white box), and an RS-rich domain (AA 117-221; dark gray box). The 4 different mutation types and their mutation localization are indicated in red. The mutation frequency is listed and also shown as black triangles above each mutation type. Each triangle is representing 1 mutated case. From top to bottom, Pro95 missense mutations, p.Pro95_Arg102del;Pro107His, p.Arg86_Gly93dup, and p.Arg94_Pro95insArg alterations are depicted.
Schematic overview of SRSF2 protein organization, mutation type, and mutation frequency. SRSF2 consists of an RRM (AA 14-92; light gray box), a linker region (white box), and an RS-rich domain (AA 117-221; dark gray box). The 4 different mutation types and their mutation localization are indicated in red. The mutation frequency is listed and also shown as black triangles above each mutation type. Each triangle is representing 1 mutated case. From top to bottom, Pro95 missense mutations, p.Pro95_Arg102del;Pro107His, p.Arg86_Gly93dup, and p.Arg94_Pro95insArg alterations are depicted.
In silico analyses
To estimate the damaging character of these specific missense mutations at Pro95, we used SIFT (http://sift.jcvi.org), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/index.shtml), and MutationTaster (www.mutationtaster.org) online analysis tools. The straightforward physical and comparative considerations showed predominantly a damaging character for all missense mutations leading to AA exchanges at position Pro95 (see supplemental Methods).
To gain insights into the extent by which other SRSF2 mutations might alter the protein folding and therefore the protein function, we generated and compared structural models of SRSF2wt and SRSF2mut. A crystal structure of the SRSF2 protein is only available for the RRM domain, and a complete structure for any of the SR proteins has not yet been achieved. To evaluate any altering character of the 3 novel mutation types p.[Pro95_Arg102del;Pro107His], p.Arg86_Gly93dup, and p.Arg94_Pro95insArg on the protein structure, we used the Robetta server (http://robetta.bakerlab.org)35 to calculate a complete structural model of the wild-type SRSF2 protein and the different mutant SRSF2 proteins (Figure 2A).
In silico analyses of the structural models of the SRSF2 in/del mutations. The structural changes of the in/del mutations were calculated with the Robetta server algorithm (http://robetta.bakerlab.org).35 The calculated model for the complete SRSF2wt protein (white structure) is depicted (A), the mutational hotspot Pro95 is marked in red. In addition, the enlargement shows the structure of AA 61-129 of the calculated models: p.[Pro95_Arg102del;Pro107His]; p.Arg86_Gly93dup; and p.Arg94_Pro95insArg. The Cα-Cα distance measurement of the corresponding AA of SRSF2wt to SRSF2mut is shown exemplarily for Pro95. All Å values of the Cα-Cα distance measurements for AA 88-99 are given (B).
In silico analyses of the structural models of the SRSF2 in/del mutations. The structural changes of the in/del mutations were calculated with the Robetta server algorithm (http://robetta.bakerlab.org).35 The calculated model for the complete SRSF2wt protein (white structure) is depicted (A), the mutational hotspot Pro95 is marked in red. In addition, the enlargement shows the structure of AA 61-129 of the calculated models: p.[Pro95_Arg102del;Pro107His]; p.Arg86_Gly93dup; and p.Arg94_Pro95insArg. The Cα-Cα distance measurement of the corresponding AA of SRSF2wt to SRSF2mut is shown exemplarily for Pro95. All Å values of the Cα-Cα distance measurements for AA 88-99 are given (B).
The differences between these models were determined by calculating the Cα-Cα distances between the 2 corresponding amino acids of SRSF2wt and mutant SRSF2 for AA 88-99. This area covers the mutation hotspot Pro95 and represents the linker sequence (AA 92-117) and, therefore, reflects the proper folding of the 2 functional domains (RRM and RS) relative to each other. The 3 analyzed novel mutations all found different distances relative to SRSF2wt, summarized in a table in Figure 2B. The 3-bp duplication showed the smallest divergence to the reference model with a distance range of 0.4-6.3 Å. The 24-bp deletion and the 24-bp insertion models show greater differences with distances ranging from 0.2 to 20.1 Å and 0.5 to 22.7 Å, respectively. Because only 1 AA is changed by the missense mutations, the models for the missense mutations show only slight divergences, being congruent with the wt SRSF2 model (Cα-Cα distances and a more detailed report about the whole procedure are given in the supplemental Methods). These data show that all calculated models very well fit the known crystal structure of the RRM domain up to AA 92, and larger changes appear within the mutated linker sequences.
Taken together, the in silico analyses indicate that the linker sequence, particularly AA 95, probably has a relevant effect on protein structure.
Correlation of SRSF2 with karyotype
As shown before, the majority of patients had a normal karyotype (71%; 190 of 269), whereas 29% (79 of 269) showed an aberrant karyotype. Within the aberrant karyotype group, the most frequent abnormalities were a loss of the Y-chromosome, aberrations of chromosome 7, and trisomy 8. Therefore, we correlated SRSF2mut with both a normal or aberrant karyotype and with subgroups exhibiting the respective chromosomal changes. These analyses found a normal karyotype in 81% of the SRSF2mut cases. Stated differently, in the group with a normal karyotype, 53% (101 of 190) had a SRSF2 mutation, whereas only 30% (24 of 79) were SRSF2mut in the aberrant karyotype group (P = .001; Table 1; Figure 3). Therefore, SRSF2mut correlated significantly with the low-risk group (composed of normal karyotype and loss of the Y-chromosome)4 compared with the intermediate-risk group (103 of 203, 51% vs 8/27, 30%; P = .043). No correlation of SRSF2mut was noted for the subcohorts with either loss of the Y-chromosome, chromosome 7 aberrations, or trisomy 8.
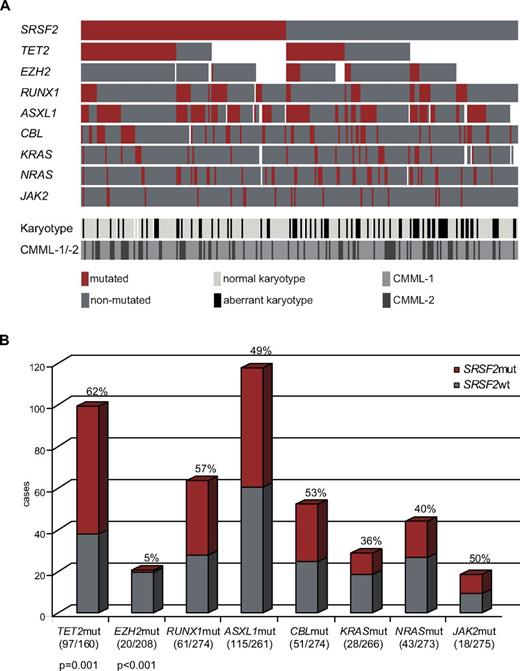
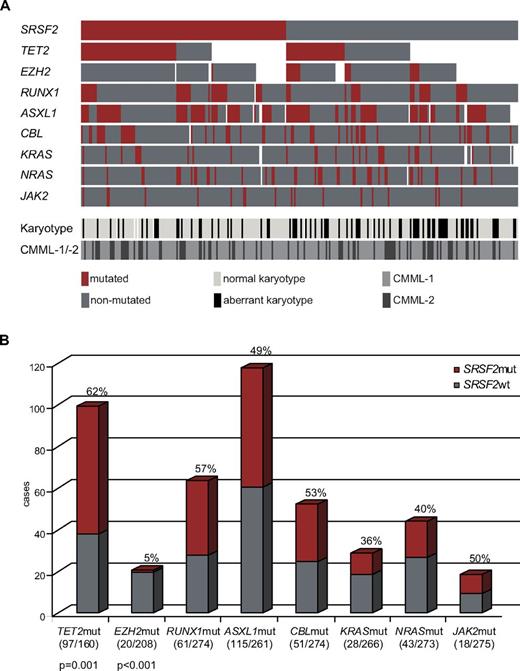
Alignment of gene mutations, karyotype information, and CMML category for 275 patients. (A) Each column represents 1 of the 275 analyzed samples. Analyses of 9 investigated genes, the karyotype, and CMML category-1 or -2 are depicted by colored bars. Red bar indicates mutated gene; dark gray bar, nonmutated gene; white bar, no data available; light-gray bar, normal karyotype; black bar, aberrant karyotype; gray bar, CMML-1; and anthracite bar, CMML-2. (B) Concomitant events of SRSF2 with other mutations are also shown as a bar chart. The gray part represents SRSF2wt, the red one SRSF2mut within the analyzed subcohorts. SRSF2mut frequencies and significances (P values) are denoted; numbers of mutated/analyzed cases of the subcohorts are given in parentheses below the bars..
Alignment of gene mutations, karyotype information, and CMML category for 275 patients. (A) Each column represents 1 of the 275 analyzed samples. Analyses of 9 investigated genes, the karyotype, and CMML category-1 or -2 are depicted by colored bars. Red bar indicates mutated gene; dark gray bar, nonmutated gene; white bar, no data available; light-gray bar, normal karyotype; black bar, aberrant karyotype; gray bar, CMML-1; and anthracite bar, CMML-2. (B) Concomitant events of SRSF2 with other mutations are also shown as a bar chart. The gray part represents SRSF2wt, the red one SRSF2mut within the analyzed subcohorts. SRSF2mut frequencies and significances (P values) are denoted; numbers of mutated/analyzed cases of the subcohorts are given in parentheses below the bars..
Correlation of SRSF2 with biologic parameters
Mutations in SRSF2 correlated with higher age (73.6 years vs 71.5 years in the SRSF2wt cases; P = .011) and higher hemoglobin levels (11.3 vs 10.2 g/dL in the SRSF2wt cases; P = .006), whereas white blood cell (WBC) and platelet counts were not different. No correlations were observed between cases with SRSF2mut and the CMML categories 1 and 2 or sex (Table 1). There was also no significant correlation of SRSF2mut with other morphologic features (supplemental Methods), any MDA risk category, or proliferative CMML (WBC counts > 13 000/μL) and dysplastic CMML (WBC counts < 13 000/μL).
Coincidence of SRSF2 with other mutations
We further investigated our CMML cohort for mutations in genes that have been described to be relevant in CMML. ASXL1, CBL, EZH2, KRAS, NRAS, IDH1/2, JAK2V617F, RUNX1, SF3B1, TET2, and U2AF1 were analyzed in large fractions of the 275 cases (Table 1). Comparison of the mutation frequencies of these genes showed that SRSF2 is the second most frequently mutated gene in this cohort (47%; 129 of 275) after TET2 (61%; 97 of 160), followed by ASXL1 (44%; 115 of 261), RUNX1 (22%; 61 of 274), CBL (19%; 51 of 274), NRAS (16%; 43 of 273), KRAS (11%; 28 of 266), EZH2 (10%; 20 of 208), and JAK2 (7%; 18 of 275). The mutation frequencies and associations are shown in Table 1 and Figure 3, respectively. Mutations in IDH1/2, U2AF1, and SF3B1 occurred in ≤ 5% of patients and are therefore depicted in supplemental Figure 3.
Analyses of coincidences showed that SRSF2 mutations were nearly mutually exclusive of EZH2 mutations. Of the 20 cases with an EZH2 mutation only 1 had a SRSF2 mutation. In counter-distinction, in the 208 cases with wt EZH2, SRSF2 was mutated in 106 samples (56%; P < .001). In contrast, a high coincidence of SRSF2 mutations occurred with TET2 mutations as 62% (60 of 97) of the samples with TET2mut had a SRSF2 mutation; whereas in the TET2wt group, only 35% (22 or 63) also carried a mutation in SRSF2 (P = .001). For associations with all the other genes, no specific associations were observed (Figure 3B). In a further analysis the coincidences of SRSF2mut with any other gene mutation were analyzed separately for CMML-1 and CMML-2 cases. Both groups reflect the same associations as observed in the total cohort (supplemental Figure 4).
Comprehensive analysis of gene mutations
In a subset of 148 cases of the cohort, the mutational status data of 9 genes were available (SRSF2, ASXL1, CBL, EZH2, JAK2, KRAS, NRAS, RUNX1, and TET2). Overall, 93% (137 of 148) of the samples had at least 1 mutation in any of these genes, whereas only 7% (11 of 148) showed no molecular mutation. Eight of these 11 patients without mutation had a normal karyotype; 3 patients carried an aberrant karyotype. This consequently leads to a combined detection rate of alterations in 140 of 148 patients with CMML (95%) having cytogenetic and/or molecular genetic aberrations. Twelve percent (18 of 148) showed mutations in 1 gene; but in none of these 18 cases did a sole mutation of either SRSF2 or RUNX1 occur. Most of the cases had simultaneous mutations in 2 (33%; 49 of 148) or 3 (28%; 42 of 148) genes. In cases with mutations involving 2 genes, 1 of the 2 mutated genes was SRSF2 in 49% (24 of 49) of the samples. In these cases the mutational load of SRSF2mut was equal or beneath the mutational load of the second mutated gene. Four mutations occurred in 22 of 148 cases (15%). In only 5 patients mutations in 5 genes were observed (5 of 148; 3%), 1 patient carried mutations in 7 genes (1 of 148; 1%).
Effect of SRSF2 mutation on clinical outcome
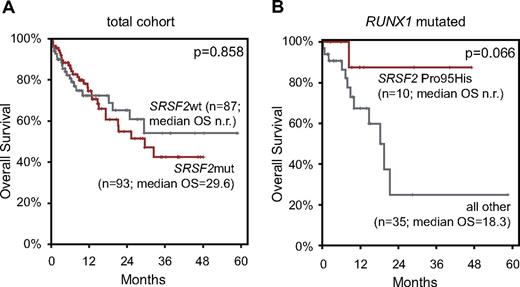
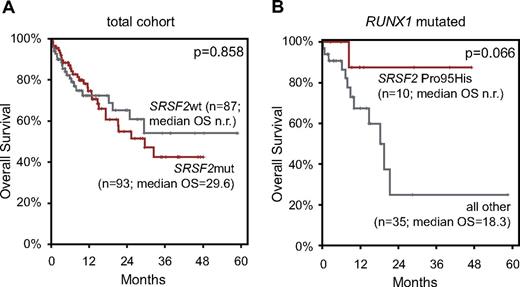
Follow-up data were available in 180 cases (median follow-up, 12 months; median OS, 29.6 months). This cohort comprised 117 CMML-1 (65%) and 63 CMML-2 (35%) cases, and 93 patients had mutations in SRSF2 (52%). Calculation of the OS for prognostic relevance of ASXL1, EZH2, TET2, and RUNX1 mutations in the total CMML cohort found an adverse effect of ASXL1mut compared with ASXL1wt (median OS, 17.3 months vs not reached; P = .001) and a slightly adverse effect of EZH2mut relative to EZH2wt (median OS, 18.3 vs 29.3 months; P = .073). TET2 and RUNX1 mutations showed no effect on OS (supplemental Figure 5).
Finally, the influence of SRSF2 mutation on survival was analyzed. In the total cohort, no effect of SRSF2 mutations on OS was observed (Figure 4A). Because of the high coincidence of SRSF2 mutations with TET2 mutations and the prognostic relevance of RUNX1 and ASXL1 alterations in MDS and CMML, respectively, we additionally analyzed these specific subcohorts, resulting in no statistically significant differences. Further, the 3 most frequently appearing missense mutations (Pro95His, Pro95Leu, and Pro95Arg) were analyzed separately. The OS curve of Pro95His-mutated cases showed a slightly better course compared with the wt SRSF2 cases, whereas the OS of Pro95Leu and Pro95Arg was slightly shorter than of the wt (see supplemental Figure 6). On the basis of these finding, we calculated the prognostic relevance of Pro95His separately in the above-mentioned subcohorts: TET2mut, RUNX1mut, and ASXL1mut. Pro95His tends to have a favorable effect on OS in the RUNX1mut group compared with other SRSF2mut or SRSF2wt cases (median OS, not reached vs 18.3 months; P = .066; Figure 4B). SRSF2mut had no influence on OS within any of the cytogenetic risk categories or MDA risk groups.
OS by Kaplan-Meier analyses of patients with CMML according to SRSF2 mutations. (A) OS of patients with SRSF2mut did not significantly differ from patients with SRSF2wt. (B) OS of patients positive for SRSF2 Pro95His compared with patients with all other SRSF2 mutations and SRSF2wt (= all other) within the RUNX1-mutated subcohort showed a favorable trend. OS is indicated in months and was compared with the 2-sided log-rank test. P values are denoted in each graph, respectively.
OS by Kaplan-Meier analyses of patients with CMML according to SRSF2 mutations. (A) OS of patients with SRSF2mut did not significantly differ from patients with SRSF2wt. (B) OS of patients positive for SRSF2 Pro95His compared with patients with all other SRSF2 mutations and SRSF2wt (= all other) within the RUNX1-mutated subcohort showed a favorable trend. OS is indicated in months and was compared with the 2-sided log-rank test. P values are denoted in each graph, respectively.
Discussion
A number of molecular targets have been identified that are frequently mutated in MDS or MDS/MPN. Thereby, some cellular pathways became apparent that are affected by mutations of several genes, including tyrosine kinase signaling and epigenetic regulation.5,6,8,20 Recently, components of the splicing machinery were found to be frequently mutated in MDS, including mutations in U2AF1, ZRSR2, SF3B1, and SRSF2.7 All of these factors are involved in 3′-splice site recognition of pre-mRNA, inducing abnormal RNA splicing.
In the present study, we analyzed 275 patients with CMML for mutations in SRSF2 and found a high frequency of mutations (47%). This frequency is even higher than the 28% that is described in the primary publication of Yoshida et al.7 This difference in frequencies may be caused by ethnic differences of the 2 cohorts, more stringent patient selection and diagnostic procedures (using in all cases nonspecific esterase for calculation of monocytes), or partially by methodologic differences. For example, the next-generation short read sequencing platform that was used in the previous study may have missed the in/del mutations. SRSF2, therefore, belongs to the most frequently mutated genes in CMML together with TET2 and ASXL1, with incidences of 61% and 44%, respectively, which is comparable with the frequencies of 44%-50%6,22 and 49%20 in previous studies.
Of note, all other results of our mutational screening were in line with published data. RUNX1 was mutated in 22% of the cases, which is in the range of findings reported by Kohlmann et al,6 and Gelsi-Boyer et al,38 with frequencies of 9%, and 30%, respectively. Likewise, CBL was mutated in 19% of the cases, and therefore in the range of 13% and 22% reported by Grand et al32 and Kohlmann et al,6 respectively. In this article with our enlarged cohort we also confirmed the mutation frequency of RAS gene mutations of 30%, also observed by Kohlmann et al6 (16% for NRAS and 11% for KRAS). Grossmann et al found a mutation frequency of 11% for EZH2,5 which was confirmed with 10%. Levine et al noted a mutation frequency of 8% for JAK2,39 which is in line with the 7% mutated cases observed in this study. IDH1/2 showed a mutation frequency of 5%,5 being in line with 4% presented in Jankowska et al.8
The cytogenetic risk stratification suggested by Such et al could not be confirmed in this cohort by Kaplan-Meier analysis,4 which may be because of small case numbers for the intermediate- (n = 18) and low-risk (n = 27) categories. The median OS for the low- and intermediate-risk groups were not reached, and the median OS for the high-risk group was 21.1 months, but there was no statistically significant difference between the 3 cohorts. This is also true for the MDA risk stratification, whereby the case numbers were even smaller (low, n = 7; intermediate-1, n = 8; intermediate-2, n = 17; high, n = 8). The median OS for the low and intermediate-1 risk groups were not reached and was 11.6 months for the intermediate-2 and 17.3 months for the high-risk groups.
For functional insights of the SRSF2 mutations various computational analyses were performed. Because a crystal structure of SR proteins is not available, bioinformatic tools were used to predict the character of the missense mutations and to generate SRSF2-structural models that were based on the amino acid sequence. All missense mutations of Pro95 in this study were predicted to be damaging. Recently, Daubner et al analyzed the RNA binding mode of SRSF2 and indicated that Pro95 forms extensive contact with RNA.40 In addition, the 3 newly described mutations with deletions and insertions are suggestive of being even more deleterious. Comparison of the calculated models indicated that the mutations affected the linker sequence. Therefore, the topography of the 2 domains (RRM and RS) might have changed as a result of an altered number or structure of the amino acid. Considering the fact that no frameshift or nonsense mutations occurred, the protein probably retains both structural integrity and any other modified function.
SRSF2 belongs to the SR protein family and is therefore a splicing factor involved in alternative splicing (reviewed in Long and Caceres25 and Shepard and Hertel26 ). Alternative splicing is an essential process by which eukaryotes generate high protein diversity from a single gene through the selective joining of different exons. More than 60% of human genes have been estimated to be alternatively spliced,41 indicating that regulation of alternative splicing is an important event. Mutations in both the nucleotide sequence of splicing regulatory elements and the components of the cellular splicing machinery can result in aberrant splicing. In addition, aberrant splicing has been found to be associated with various diseases, including cancer.42,43 Many cancer-related genes are regulated by alternative splicing, and changes in the splicing pattern appear to be unique to the malignant state.44,45 Daubner et al report that mutations of SRSF2 affecting the RRM also affect the function, showing a decreased splicing activity of the protein.40 More recently, Makishima et al showed that SRSF2mut leads to defective splicing of the RUNX1 gene.46 Moreover, miss-expression of SR proteins changes the alternative splicing pattern and is associated with the development of cancer. Increased expression of SR proteins correlates with cancer progression, as was shown for SRSF2 in ovarian cancers.47 However, depletion of SRSF2 in the thymus of a mouse model changed the alternative splicing of CD45, causing a defect in T-cell maturation.48 Lareau et al reported that SRSF2 directs the splicing of its own transcripts and autoregulates its own expression by coupling alternative splicing with RNA decay.49 Recent reports indicate further functions of SRSF2 in transcription, promoting RNA Pol II elongation, genome stability, and cell-cycle progression (reviewed in Long and Caceres25 and Zhong et al50 ). Taken together, mutations in SRSF2, although occurring in a region without any obvious functional domain, may cause changes in protein function or expression levels, both possibly contributing to a change of alternative splicing patterns, leading to developmental defects and the onset of cancer.
SRSF2 mutations frequently overlapped with other mutations in our cohort of 275 patients with CMML. Only mutations of EZH2 did not overlap, pointing to their mutual exclusiveness. One may speculate that this occurs because either no advantageous cooperating effect results from both proteins being altered or concomitant mutations of both proteins is lethal for the cell. Overall, in 18 cases only 1 mutation was detected and this was never SRSF2. Thus, SRSF2 never occurs as a sole mutation, either indicating that SRSF2 mutations are not early events in the pathogenesis of CMML or that a sole mutation in SRSF2 results in no clinical manifestation. This is further supported because the mutational load of SRSF2mut was always equal or below the mutational load of the second mutated gene in cases with only 2 mutations. By contrast, SRSF2 is frequently mutated in cases with either 2 or 3 mutations. SRSF2 mutations may result in a dysfunction of the protein that affects transcriptional elongation and therefore genome stability. Depletion of SRSF2 has been reported to trigger overwhelming double-strand breaks (reviewed in Zhong et al50 ).
Mutations in SRSF2 were highly associated with TET2 mutations, a protein converting 5-methyl-cytosin to 5-hydroxymethyl-cytosin. Depletion of TET2 in bone marrow progenitor cells promotes an expansion of monocyte/macrophage cells,51 indicating that loss of function can promote clonal expansion of mutant cells. Addressing the WBC count in cases with TET2wt + SRSF2wt (n = 32) showed a mean of 16 036 cells/μL, whereas TET2mut + SRSF2wt cases (n = 33) showed 31 112 WBC/μL (P = .044). SRSF2mut seems to antagonize this leukocytosis, mostly by monocytosis, because the mean WBC count was 16 864/μL in cases with TET2mut + SRSF2mut (n = 57; TET2mut + SRSF2mut vs TET2wt + SRSF2wt; P = .047). As mentioned earlier, SRSF2 depletion has been reported to cause genome instability by triggering double-strand breaks, which induced the S phase checkpoint and ended in cell cycle arrest or apoptosis (reviewed in Zhong et al50 ). Furthermore, SRSF2 mutation is correlated with higher hemoglobin levels. Thus, patients with SRSF2mut show a less pronounced leuko/monocytosis in the presence of a concomitant TET2 mutation and have a less pronounced anemia, both indicating a better state of health.
The median OS of our cohort is 29.6 months, indicating that the outcome of our cohort is somehow better than in other datasets published (eg, Onida et al3 ). This may be because 60% of the patients sent for diagnosis to our institution are referred from outpatient units and hematologist practices at first suspect of CMML and thus are diagnosed very early. Many of our patients were not treated upfront but followed a watch-and-wait-strategy. This may in part explain the differences in the survival curves in comparison with other studies published from centers to which the patients were referred to receive treatment, including enrollment into clinical trials. The mutational status of SRSF2 did not affect OS, although the median OS was not reached in SRSF2wt cases in contrast to 29.6 months in cases with a mutated SRSF2 (P = .858). In RUNX1-mutated cases the addition of a SRSF2 mutation prolonged the OS. Analyzing the most frequently occurring SRSF2 missense mutations (Pro95His, Pro95Leu, and Pro95Arg) separately indicated that CMML with a Pro95His showed a better outcome than the other 2 frequent mutations as well as the wild-type SRSF2, in the RUNX1, TET2, and ASXL1 mutated groups. This goes in line with the idea that a SRSF2 mutation, especially Pro95His, affects protein function; this may result in a favorable effect in cases with concomitant (adverse) mutation, possibly because of inhibition of cell cycle progression.
In summary, SRSF2 mutations are common in CMML and seem to have a deleterious effect on protein structure and function. This may on the one hand result in promoting further gene mutations and therefore disease progression. On the other hand, it could have a favorable effect on the OS of patients with an additional (adverse) mutation. SRSF2mut further correlated with a normal karyotype and confined the cytogenetic categories low and intermediate. SRSF2, therefore, represents a novel molecular marker that is helpful for diagnosis of CMML or suspected CMML and for further genetic characterization of this disease. A possible positive prognostic effect in cases with other, partially adverse mutations (RUNX1, TET2, and ASXL1), that was suggested based on our results has to be validated in further independent studies. Of note, based on this data, overall 93% of patients with CMML in the present cohort carried at least 1 mutated gene. However, cases are still found without any detectable genetic defect, warranting further efforts to identify new genetic aberrations that are essential to better understand the molecular pathology of this disease.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: M.M. investigated the molecular mutations of SRSF2 and ASXL1, analyzed the data, and wrote the manuscript; A.R. made the bioinformatic analyses; T.H. was responsible for cytomorphologic analysis and was involved in the collection of clinical data; C.E., F.D., V.G., and A.K. contributed to molecular analyses of the ASXL1, CBL, EZH2, JAK2V617F, KRAS, NRAS, RUNX1, and TET2 mutations; T.A. collected and documented clinical data and compiled statistical analyses; K.Y., S.O., and H.P.K. originally detected SRSF2 gene mutations and shared unpublished data; W.K. was responsible for immunophenotyping and was involved in statistical analyses; C.H. was responsible for chromosome banding analysis; S.S. was the principle investigator of the study and wrote the manuscript. All authors read and contributed to the final version of the manuscript.
Conflict-of-interest disclosure: Several of the authors (T.H., W.K., C.H., and S.S.) are part owners of the MLL Munich Leukemia Laboratory. Several of the authors (M.M., A.R., C.E., F.D., A.K., V.G., and T.A.) are employed by the MLL Munich Leukemia Laboratory. K.Y., S.O., and H.P.K. declare no competing financial interests.
Correspondence: Susanne Schnittger, MLL Munich Leukemia Laboratory, Max-Lebsche-Platz 31, 81377 Munich, Germany; e-mail: susanne.schnittger@mll.com.

![Figure 2. In silico analyses of the structural models of the SRSF2 in/del mutations. The structural changes of the in/del mutations were calculated with the Robetta server algorithm (http://robetta.bakerlab.org).35 The calculated model for the complete SRSF2wt protein (white structure) is depicted (A), the mutational hotspot Pro95 is marked in red. In addition, the enlargement shows the structure of AA 61-129 of the calculated models: p.[Pro95_Arg102del;Pro107His]; p.Arg86_Gly93dup; and p.Arg94_Pro95insArg. The Cα-Cα distance measurement of the corresponding AA of SRSF2wt to SRSF2mut is shown exemplarily for Pro95. All Å values of the Cα-Cα distance measurements for AA 88-99 are given (B).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/120/15/10.1182_blood-2012-01-404863/4/m_zh89991297420002.jpeg?Expires=1766500273&Signature=yjuxNh-IYxsqXIt4mrwOU7pK7ekUMkWfEdC6YBKwErj5RMNDNT21~ZpGsbOoXd8SWgNqHS0kebD~yM5uMmz4VJt3gXZq6Kja0bFjq-V82kuy2W4fYZHzuuGZnnCHak5IJTJ8R-5gF8FGAuRG3dFRIEEeStczQ2hGns1jDM4sNsPzoFJNvRvYpwFLh3jYNYQQewOUKEPsiCDm4WoIELqWnmxxDiZ1JVCtZ2qrSK06cM1QZCODMACOMnFrcFs12drzx3DPSPYTFMV1venq9c3igoGd5v~UDMbWsEV9l3Xmy4yd0kp9it~YEwxBBSoeNm6jsipnS5Vfg4sBZjEB80~7xw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 2. In silico analyses of the structural models of the SRSF2 in/del mutations. The structural changes of the in/del mutations were calculated with the Robetta server algorithm (http://robetta.bakerlab.org).35 The calculated model for the complete SRSF2wt protein (white structure) is depicted (A), the mutational hotspot Pro95 is marked in red. In addition, the enlargement shows the structure of AA 61-129 of the calculated models: p.[Pro95_Arg102del;Pro107His]; p.Arg86_Gly93dup; and p.Arg94_Pro95insArg. The Cα-Cα distance measurement of the corresponding AA of SRSF2wt to SRSF2mut is shown exemplarily for Pro95. All Å values of the Cα-Cα distance measurements for AA 88-99 are given (B).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/120/15/10.1182_blood-2012-01-404863/4/m_zh89991297420002.jpeg?Expires=1766972269&Signature=NL~rJFbTVqsr8FiuAWOBWM6nkHY8KDz4jIHraoUUmCeYM5t4digwgtV-V8TEXpaDLxnjMKOI--pbeqZOt0cDSWhHbDU4domabM89fbF95lTwoNKDL0irzskNJm40fbgMVEZfhwzHi3dnPyYLshn8JnfYqe8F5EQEwtX-f-PaOlGh0mg5KXhRSa4-5xCMGKIppzMYREAWfoWgYICxn5DF2TD3xIak0utMRw8XNl1SXKoaTCkIv64gruP3Fe2RWSyPNgKXLSKB09HrVxodVschN206o1MtOJpelDnnHgCM9CCGedWei1K2NbrC0OUlL8QV7qLlV5-OIKGPKen0HBoZOA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)