Abstract
Missense mutations that reduce or abrogate myeloid cell expression of the F-BAR domain protein, proline serine threonine phosphatase-interacting protein 2 (PSTPIP2), lead to autoinflammatory disease involving extramedullary hematopoiesis, skin and bone lesions. However, little is known about how PSTPIP2 regulates osteoclast development. Here we examined how PSTPIP2 deficiency causes osteopenia and bone lesions, using the mouse PSTPIP2 mutations, cmo, which fails to express PSTPIP2 and Lupo, in which PSTPIP2 is dysfunctional. In both models, serum levels of the pro-osteoclastogenic factor, MIP-1α, were elevated and CSF-1 receptor (CSF-1R)–dependent production of MIP-1α by macrophages was increased. Treatment of cmo mice with a dual specificity CSF-1R and c-Kit inhibitor, PLX3397, decreased circulating MIP-1α and ameliorated the extramedullary hematopoiesis, inflammation, and osteopenia, demonstrating that aberrant myelopoiesis drives disease. Purified osteoclast precursors from PSTPIP2-deficient mice exhibit increased osteoclastogenesis in vitro and were used to probe the structural requirements for PSTPIP2 suppression of osteoclast development. PSTPIP2 tyrosine phosphorylation and a functional F-BAR domain were essential for PSTPIP2 inhibition of TRAP expression and osteoclast precursor fusion, whereas interaction with PEST-type phosphatases was only required for suppression of TRAP expression. Thus, PSTPIP2 acts as a negative feedback regulator of CSF-1R signaling to suppress inflammation and osteoclastogenesis.
Introduction
The combination of chronic immune activation and musculoskeletal tissue damage is the hallmark of rheumatic diseases.1 Osteolytic lesions coupled with skin and/or joint inflammation occur in several rheumatic conditions, such as rheumatoid arthritis, psoriatic arthritis, and chronic recurrent multifocal osteomyelitis (CRMO).1,2 Thus, an understanding of the pathophysiologic mechanisms underlying rheumatic disease requires the identification of the molecular pathways that simultaneously regulate inflammation and bone homeostasis.
Osteoclasts are bone-resorbing multinucleated giant cells of myeloid origin. Receptor activator of nuclear factor κB ligand (RANKL) and colony stimulating factor-1 (CSF-1) are necessary and sufficient for osteoclast differentiation from monocytic precursor cells in vivo and in vitro.3-5 CSF-1 modulates multiple steps of osteoclastogenesis, including proliferation of mononuclear OC precursors (OCP), their differentiation and their fusion. In synergy with RANKL, CSF-1 also stimulates the expression of several osteoclast-specific genes including RANK, components of RANK signaling pathways and tartrate-resistant acid phosphatase (TRAP).6-9
Proline serine threonine phosphatase-interacting protein 2 (PSTPIP2), also known as macrophage F-actin–associated and tyrosine phosphorylated protein (MAYP), is a Fes CIP4 homology domain (FCH) and Bin/Amphiphysin/Rvs (BAR; F-BAR) protein, predominantly expressed in the myeloid lineage.10 It is rapidly tyrosine phosphorylated after activation of CSF-1 receptor (CSF-1R),10-14 and exhibits reduced phosphorylation in mast cells in which c-Kit is inhibited.12 The mouse Pstpip2 missense mutations, chronic multifocal osteomyelitis (cmo), L98P (Pstpip2cmo/cmo), and Lupo I282N (Pstpip2Lupo/Lupo) lead to similar autoinflammatory diseases, both characterized by splenomegaly, skin necrosis, and aseptic osteomyelitis. These diseases resemble CRMO and psoriatic arthritis in man.11,14-17 The mutations result in complete (cmo) or partial (Lupo) PSTPIP2 protein deficiency, indicating that PSTPIP2 has anti-inflammatory activity.11,14 Histologic studies of the cmo mice showed osteoclast-mediated bone resorption at sites of inflammation in caudal vertebrae,15,17 and cultured cmo bone marrow cells exhibited increased vitamin D3–induced osteoclastogenic responses.17 However, the molecular bases of these phenotypes were not elucidated.
In this study, we show that, in addition to the bone erosive disease, PSTPIP2 deficiency leads to generalized osteopenia and CSF-1R–dependent elevation of osteoclast precursors and of serum MIP-1α. Absence of PSTPIP2 causes a cell autonomous defect favoring osteoclastogenesis from multipotent myeloid precursors. In addition, we demonstrate that several distinct molecular interactions of PSTPIP2 are required for suppression of osteoclast differentiation at different stages. Although CSF-1 and RANKL positively regulate osteoclastogenesis,6-9 our results demonstrate that CSF-1R–regulated PSTPIP2 tyrosine phosphorylation is required for suppression of osteoclastogenesis, indicating that PSTPIP2 normally plays a negative feedback role.
Methods
Antibodies and reagents
The dual specificity inhibitor, PLX3397, was a gift from Plexxikon. RANKL was purchased from Cell Sciences. Anti-CD117–FITC, anti-CD11b–APC, anti-CD16/CD32–PE, anti-Ly6C–FITC, anti-CD11c–FITC, anti-CD48–FITC, anti-CD34–FITC, anti-CD150–PE, and streptavidin-PE were from BD Pharmingen. Pacific Blue anti–Sca-1, anti-CD49b–APC, anti-Ly6G–PerCP, and anti-CD3–FITC were from BioLegend. Anti-B220–PE-Cy5, anti–CD4-PE–Cy5, anti–CD19-PE–Cy5, anti-CD8–PE-Cy5, anti-CD127PE, anti-CD117–APC, biotinylated-AFS98, and anti-Thy1.1–FITC were from eBioscience. CSF-1 was a gift from Chiron Corporation. Unless otherwise specified, all other reagents were purchased from Sigma-Aldrich.
Mice and genotyping
Pstpip2cmo/cmo BALB/cAnPt and wild-type (WT) BALB/cByJ mice (The Jackson Laboratory) and Pstpip2Lupo/Lupo C3HeB/FeJ and WT C3HeB/FeJ mice (Ingenium Pharmaceuticals) were maintained under specific pathogen-free conditions in a barrier facility of the Albert Einstein College of Medicine Animal Institute, which approved the mouse breeding and study protocols. In addition, this study was conducted in accordance with the Declaration of Helsinki. Pstpip2 mutation genotyping was performed by PCR amplification and sequencing as described.11,14
Treatment with PLX3397 and scoring of inflammation
Treatment with PLX3397 or control chow was initiated at 5 weeks of age, before the onset of clinical disease. Inflammation was scored weekly by visual examination using the following criteria: (1) Skin for ears: 1 point for each of the following: erythema, edema, tissue hardening, or necrosis. Score doubles for bilateral symptoms. For body hair loss: localized, 1 point; general, 2 points. (2) Paws: 1 point for each of the following: bulbous toe, local signs of erythema, edema, tissue hardening, or necrosis. Score doubles if symptoms are generalized or bilateral. (3) Tails: 1 point for each tail kink and 1 point for swelling or redness.
Micro-computed tomography
After serial fixation in 4% phosphate-buffered formaldehyde and 70% ethanol, bones were scanned by high resolution micro-CT. Imaging was performed using vivaCT 40 with a voxel size of 10.5 μm (see Figure 1), and with μCT 35 (both Scanco Medical) with a voxel size of 7 μm (see Figure 3). Structural parameters were calculated using Scanco Medical Version 6 software on an area extending 2.1 mm from the metaphysis for trabecular bone and 0.6 mm at the femoral midshaft. Analysis was performed using segmentation values of 0.8/1/375 for cortical data and 0.8/1/250 and 0.8/1/275 for trabecular data in Figures 1 and 3, respectively. Paws, tail, and/or spine were imaged with vivaCT40, voxel size 15 μm, segmentation values of 0.7/1/425 (see Figure 1), or with μCT 35, voxel size 12.5 μm, segmentation values of 0.8/1/300 (see Figure 3).
Histologic and ultrastructural analysis
For TRAP staining, mice were anesthetized and perfused with periodate-lysine-paraformaldehide-glutaradehyde (PLPG) fixative by injection of the fixative in the heart.18 Tissues were dissected, further fixed by immersion in PLPG, and embedded in paraffin. Sections stained for TRAP and counterstained with hematoxylin were analyzed by light microscopy, using a Zeiss AxioSkop 2 (Carl Zeiss Microscopy) equipped with a Plan Neofluar objective 2.5×/0.075 NA and the images collected using Axiovision 4.1 software. For electron microscopic studies, 4-month-old mice were perfused with 2% paraformaldehyde and 2.5% glutaraldehyde in cacodylate buffer, pH 7.4. Tail vertebrae were fixed and processed as previously described.19 Sections were examined using a JEOL 1200EX transmission electron microscope (JEOL) and representative regions were photographed. Images were cropped and adjusted for brightness, contrast, and color saturation using Adobe Photoshop Elements 2.0.
Analysis of osteoclast and myeloid precursors by flow cytometry
Bone marrow cells, splenocytes and blood were collected from mice subjected to the indicated treatments. After red blood cell lysis and Fc receptor blocking, cells were labeled with anti-CD117–FITC, anti-CD11b–APC, biotinylated Rat IgG 2a anti–CSF-1R AFS98, and streptavidin-PE. Bone marrow populations containing myeloid and osteoclast precursors were identified as described,8,20 and analyzed or sorted using a BD-LSR II, or a BDFACS Aria I cell sorter (Becton Dickinson), respectively. Spectral overlaps between fluorophores corrected by electronic compensation. FACS data were analyzed using FlowJo Version 8.8.6 software (TreeStar).
Osteoclast differentiation and precursor proliferation
Osteoclasts were generated from bone marrow cells extracted from the femurs of WT or Pstpip2cmo/cmo mice, or from FACS-sorted osteoclast precursors, in triplicate cultures containing 30 ng/mL M-CSF and 100 ng/mL RANKL, as previously described.8,21 Cells were fixed at the indicated times and stained for TRAP as described elsewhere.22 Cells were examined under an Olympus CK2 inverted microscope (Olympus) equipped with an Olympus 10× objective, 0.25 NA. Digital images were acquired using a Kodak DC290 camera equipped with an Optem (1×) microscope adaptor and the Kodak camera digital software. Images were cropped and adjusted for brightness, contrast, and color balance using Photoshop Elements 2.0. TRAP activity was also determined in cultures grown in 96-well plates as described.23
Bone resorption assay
Bone marrow cells were plated on whale dentine slices (Immunodiagnostic Systems) for 6 days in the presence of 30 ng/mL CSF-1 and 100 ng/mL RANKL with a change of media at day 3. Cells were fixed in 4% paraformaldehyde for 20 minutes at room temperature and stained for TRAP. In some experiments, cells were removed by sonication in 0.25M ammonia hydroxide, the resorptive pits stained with toluidine blue and the stained dentin slices analyzed by light microscopy using a Zeiss STEMI (Carl Zeiss Microscopy) and IPLab 4.0.8 software. The quantitation of resorption was performed using ImageJ Version 1.4lo software. Images were cropped and adjusted for brightness, contrast, and color balance using Photoshop Elements 2.0. The same methods were applied to dentine slice implants.
Dentine implantation, PLX3397 treatment, and measurement of serum markers for inflammation
Dentine discs (Immunodiagnostic Systems) were incubated overnight in α-MEM with 10% FCS, 30 ng/mL CSF-1, and 100 ng/mL RANKL and implanted bilaterally in the gluteus maximus of mice under general anesthesia. Mice were euthanized at the indicated times and the discs were removed and stained for TRAP or resorption pits as described under “Bone resorption assay.” Blood was collected at the indicated times and sera were screened for cytokine production using multiplexed beadlyte mouse cytokine kits (Millipore) as described14 or using the mouse IL-6 ELISA Ready-SET-Go kit (eBioscience). Osteoclast precursors in blood, bone marrow, and spleen were analyzed by flow cytometry as described under “Analysis of osteoclast and myeloid precursors by flow cytometry.”
Generation of osteoclast precursor cell lines and retroviral reconstitution of PSTPIP2 expression
Early c-Kit+ Mac-1lo c-Fmslo osteoclast precursors were purified by FACS sorting and immortalized using a retroviral construct encoding SV40 large T antigen as described.14 PSTPIP2 mutants were prepared using a QuikChange site-directed mutagenesis kit (QIAGEN) and the presence of the desired mutations was confirmed by DNA sequencing. PSTPIP2 expression was reconstituted by MSCV-IRES-GFP retroviral transduction as described.24
Biochemical characterization of PSTPIP2 mutants
For lipid binding studies, PSTPIP2 was cloned in pGEX-6-P1, and expressed in Escherichia coli as GST-fusion protein. GST-PSTPIP2 was purified by affinity chromatography and the GST tag was removed by digestion with PreScission protease. WT PSTPIP2 and mutants were then tested for phospholipids binding ability using PIP strips (Echelon Biosciences). Cell stimulation, immunoprecipitation and Western Blotting were performed as described.24
PSTPIP2 in vitro tubulation assay and CD Spectroscopy
Folch fraction I (Sigma-Aldrich) liposomes were prepared by classic dehydration-rehydration protocols. Lipids were prepared in a 1:1 chloroform:methanol mix and dried in glass tubes under Argon gas. Liposomes were rehydrated in buffer (150mM NaCl, 20mM HEPES pH 7.4, 2.5mM DTT), sonicated, and filtered through 0.8-μm pore diameter polycarbonate filters (Whatman) to a final concentration of 1 mg/mL. Purified PSTPIP2 was mixed at 5 to 12μM with 0.5 mg/mL liposomes (final concentrations) and allowed to incubate for 30 minutes, and then adhered to glow-discharged carbon-coated TEM grids (Agar Scientific) for 1 minute. Samples were briefly dehydrated, negative stained with 2% uranyl acetate, and then briefly washed with water and imaged. All samples were imaged on a PW6010/20 EM2055 transmission electron microscope at 80 kV (Philips).
To assess protein folding, reconstituted PSTPIP2 proteins were resuspended in 5mM HEPES pH 7.4, 150mM NaCl. CD spectra were obtained using a Jobin Yvon CD6 CD spectrophotometer (Horiba).
Fluorescence light microscopy
Living COS-7 cells were cultured on glass-bottom culture dishes (MatTek) in DMEM–GlutaMAX-1 media (Invitrogen) and transfected using GeneJuice (EMD) with plasmids encoding N-terminally EGFP-tagged PSTPIP2 (Clontech). Cells were allowed to express the proteins overnight before imaging. For imaging, cells were placed in a temperature-controlled chamber heated to 37°C that was installed on a fully motorized inverted light microscope Eclipse TE-2000 (Nikon) equipped with a CSU-X1 spinning disk confocal unit (UltraVIEW VoX) and four 50-mW solid-state lasers. Living cells were imaged through a 60× lens (Plan Apochromat VC, 1.4 NA, Nikon). The system was operated through Volocity 5.0 (Improvision) software. Video images were acquired at 2 frames/second. Images were obtained with a cooled EMCCD camera (9100-02; Hamamatsu).
Statistics
Significance was tested using the 2-tailed Student t test.
Results
Diseased PSTPIP2-deficient Pstpip2cmo/cmo mice are osteopenic and have erosive bone lesions
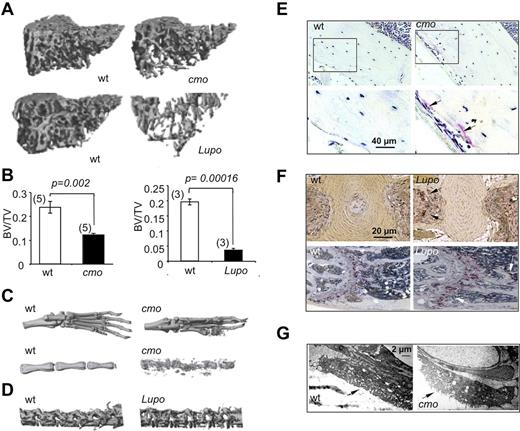
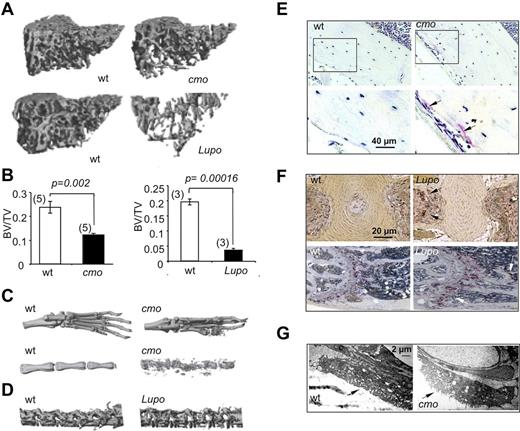
Diseases characterized by erosive arthritis frequently manifest generalized bone loss.1 To determine whether this was also a characteristic of the autoinflammatory and erosive disease seen in PSTPIP2-deficient mice, we examined trabecular and cortical bone parameters by microcomputed tomography (micro-CT). Proximal tibia trabecular bone was significantly decreased in diseased female cmo and Lupo mice, compared with age-and sex-matched WT controls (Figure 1A-B). Similar differences were seen at the distal femur and in cohorts of diseased male mice compared with WT controls (data not shown). Consistent with previous reports demonstrating osteolysis in the caudal vertebrae,15,17 representative 3D micro-CT reconstructions of paws and proximal tails of diseased cmo mice show that they developed erosive bone lesions at sites of inflammation (Figure 1C). In addition, although Lupo mice did not develop kinked tails, significant bone erosion was detected in the lumbar region of the spine (Figure 1D bottom panels). Thus, as observed in psoriatic or rheumatoid arthritis,1 the inflammatory diseases of PSTPIP2-deficient mice lead to both localized bone erosions and generalized bone loss.
Diseased PSTPIP2-deficient mice are osteopenic and have erosive bone lesions that are associated with increased osteoclast numbers and activity. (A) Representative 3D reconstructions of trabecular areas of proximal tibias from PSTPIP2-deficient 3.5-month-old cmo and 3-month-old Lupo mice (right panels) and their corresponding WT control mice (left panels). (B) Quantitative micro-CT analyses of 3-month-old female mice showing that, compared with WT controls (white bars), PSTPIP2-deficient mice (black bars) have decreased proximal tibia bone volume to total volume (BV/TV). Figures in parentheses indicate the numbers of mice in each group. (C) Representative images of paws and proximal tails from cmo and WT control mice. Erosive bone lesions are seen on micro-CT reconstructions of paws and tails of diseased cmo mice. (D) Lumbar vertebrae from Lupo mice demonstrate osteopenia. (E) Increased TRAP staining in sections of femoral cortex of cmo compared with WT controls. Bottom panels are higher magnifications of the boxed regions in the top panels. (F) TRAP staining of tail vertebrae (top panels) and proximal femurs (bottom panels) of Lupo mice and WT controls. Arrows indicate TRAP+ osteoclasts. (G) Electron micrographs of vertebral osteoclasts. Arrows indicate the ruffled border.
Diseased PSTPIP2-deficient mice are osteopenic and have erosive bone lesions that are associated with increased osteoclast numbers and activity. (A) Representative 3D reconstructions of trabecular areas of proximal tibias from PSTPIP2-deficient 3.5-month-old cmo and 3-month-old Lupo mice (right panels) and their corresponding WT control mice (left panels). (B) Quantitative micro-CT analyses of 3-month-old female mice showing that, compared with WT controls (white bars), PSTPIP2-deficient mice (black bars) have decreased proximal tibia bone volume to total volume (BV/TV). Figures in parentheses indicate the numbers of mice in each group. (C) Representative images of paws and proximal tails from cmo and WT control mice. Erosive bone lesions are seen on micro-CT reconstructions of paws and tails of diseased cmo mice. (D) Lumbar vertebrae from Lupo mice demonstrate osteopenia. (E) Increased TRAP staining in sections of femoral cortex of cmo compared with WT controls. Bottom panels are higher magnifications of the boxed regions in the top panels. (F) TRAP staining of tail vertebrae (top panels) and proximal femurs (bottom panels) of Lupo mice and WT controls. Arrows indicate TRAP+ osteoclasts. (G) Electron micrographs of vertebral osteoclasts. Arrows indicate the ruffled border.
Increased osteoclast density and osteoclast activation in cmo mice
Histologic analysis revealed increased TRAP staining in the long bones of both cmo (Figure 1E) and Lupo mice (Figure 1F), indicative of increased osteoclast formation. Furthermore, electron microscopic examination of osteoclast morphology revealed that compared with WT mice, tail vertebrae sections obtained from cmo mice contained osteoclasts exhibiting prominent ruffled borders (Figure 1G), indicative of active bone resorption.25
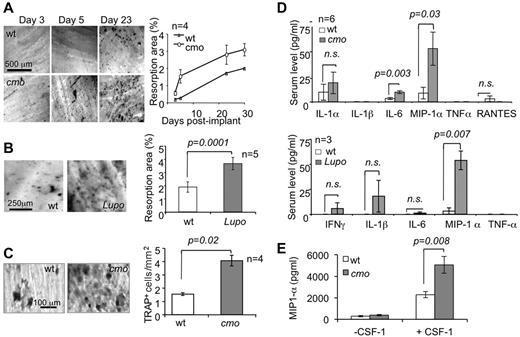
PSTPIP2-deficient mice exhibit accelerated and enhanced resorption of dentine disc implants and increased serum levels of the pro-osteoclastogenic factor MIP-1α
To test the impact of PSTPIP2-deficiency disease in mice on osteolytic responses in vivo, we performed intramuscular dentine disc implantation in PSTPIP2-deficient and WT mice. Both cmo (Figure 2A) and Lupo mice (Figure 2B) display more extensive osteolytic responses against the dentine implants than WT controls. This phenotype is associated with increased formation of TRAP+ cells on the surface of the implant (Figure 2C) and with increased circulating levels of MIP-1α, without significant elevation of other factors involved in osteoclast differentiation or osteoclast precursor mobilization, including IL-1α, IL-1β, TNF-α, or RANTES (Figure 2D). Although circulating levels of another pro-osteoclastogenic factor, IL-6, were significantly increased in cmo mice (Figure 2D), they were normal in 3-month-old Lupo mice (Figure 2D bottom panel). To confirm this, we carried out additional measurements of serum IL-6, using the more sensitive ELISA assay. There was no significant difference between serum IL-6 levels of WT and Lupo mice at either 3 months (WT 6.1 ± 2.6 pg/mL, n = 7; Lupo, 7.9 ± 2.1 pg/mL, n = 12; P = .62) or 8 months (WT 7.4 ± 3.4 pg/mL, n = 3; Lupo, 10.9 ± 2.4 pg/mL, n = 9, P = .45) of age. The significant loss of trabecular bone in 3-month-old Lupo mice (Figure 1B), in the absence of elevated IL-6 suggests that IL-6 is not a major contributor to the osteopenic phenotype of PSTPIP2-deficient mice. MIP-1α production by macrophages was dependent on CSF-1R signaling and was significantly higher in the absence of PSTPIP2 expression (Figure 2E), suggesting that PSTPIP2 negatively controls CSF-1–stimulated MIP-1α production.
PSTPIP2 deficiency promotes osteoclast development and MIP-1α production in vivo. (A) Toluidine blue staining (left panels) and quantitation (right panel) of the resorption pits on dentine implants removed at the indicated times after surgery. (B) Resorption pits (left panels) and quantitation (right panel) of dentine resorption at day 10 postimplant. (C) TRAP staining of dentine implants (left panels) and quantitation of the density (right panel) of TRAP+ cells on implants removed at day 5. (D) Serum levels of pro-osteoclastogenic factors in 6-month-old cmo (top panel) and 3-month-old Lupo (bottom panel) mice and WT controls at day 8 after surgery. (E) MIP-1α production by macrophages after a 24-hour incubation in medium with or without 120 ng/mL CSF-1. Data ± SEM; n indicates number of mice per group; and ns, not significantly different (P > .05).
PSTPIP2 deficiency promotes osteoclast development and MIP-1α production in vivo. (A) Toluidine blue staining (left panels) and quantitation (right panel) of the resorption pits on dentine implants removed at the indicated times after surgery. (B) Resorption pits (left panels) and quantitation (right panel) of dentine resorption at day 10 postimplant. (C) TRAP staining of dentine implants (left panels) and quantitation of the density (right panel) of TRAP+ cells on implants removed at day 5. (D) Serum levels of pro-osteoclastogenic factors in 6-month-old cmo (top panel) and 3-month-old Lupo (bottom panel) mice and WT controls at day 8 after surgery. (E) MIP-1α production by macrophages after a 24-hour incubation in medium with or without 120 ng/mL CSF-1. Data ± SEM; n indicates number of mice per group; and ns, not significantly different (P > .05).
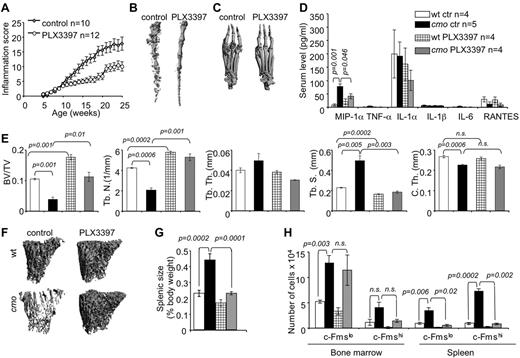
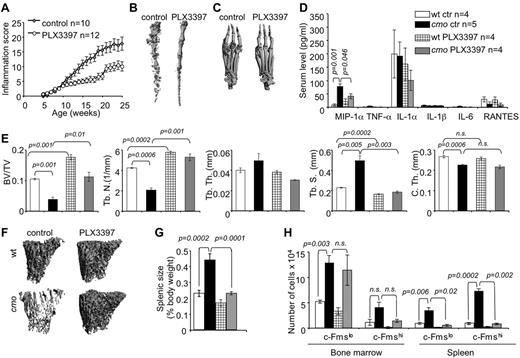
Inhibition of CSF-1R and c-Kit signaling reduces inflammation, the availability of OC precursors and attenuates disease in cmo mice
Stem cell factor (c-Kit ligand) and CSF-1 together regulate the proliferation of multipotent hematopoietic progenitor cells and their myeloid differentiation.26 Osteoclast precursors are substantially enriched among the c-Kit+ c-Fmslo Mac1lo multipotent myeloid precursor (early osteoclast precursor, EOCP) and the c-Kit+ c-Fmshi Mac1lo bipotent (monocyte/osteoclast) precursor (late osteoclast precursor, LOCP) fractions of the bone marrow.8 We observed that both cmo bone marrow and spleen contain increased numbers of these precursors (Figure 3H). To test whether CSF-1 and c-Kit–dependent expansion of OCP significantly contributes to the inflammation and osteopenia in cmo mice, we administered PLX3397, a CSF-1R/c-Kit dual-specificity inhibitor,27 to asymptomatic cmo mice and examined its effect on disease development. PLX3397 significantly attenuated autoinflammatory disease (Figure 3A), decreasing the erosive bone lesions in tails and paws (Figure 3B-C) and the levels of circulating MIP-1α (Figure 3D) in cmo mice. Moreover, PLX3397 completely abrogated the generalized trabecular bone loss seen in diseased cmo mice and also increased trabecular bone in WT mice (Figure 3E-F). However, there was no effect of PLX3397 treatment on cortical thickness in either WT or cmo mice (Figure 3E).
Inhibition of myelopoiesis using a dual c-Fms/c-Kit inhibitor attenuates inflammation and bone destruction in cmo mice. (A) Time course of disease progression in cmo mice receiving the c-Fms/c-Kit inhibitor, PLX3397 (PLX), or control chow. Treatment was started at 5 weeks of age, before the onset of clinical symptoms and continued up to ∼ 8 months of age (WT, 252 ± 18 and cmo 248 ± 19 days), at which time the mice were euthanized and analyzed. (B-C) Micro CT reconstruction of distal tails (B) and hind paws (C) of PLX-treated and control cmo mice. (D) PLX treatment decreases circulating MIP-1α in cmo mice. (E) Quantitative bone parameters of PLX-treated and control cmo and WT mice. (F) Micro CT reconstructions of proximal tibiae. (G-H) PLX treatment of cmo mice prevents splenomegaly (G) and the expansion of splenic OCP (H). Scale bars, 100 μm. Data ± SEM, n ≥ 3.
Inhibition of myelopoiesis using a dual c-Fms/c-Kit inhibitor attenuates inflammation and bone destruction in cmo mice. (A) Time course of disease progression in cmo mice receiving the c-Fms/c-Kit inhibitor, PLX3397 (PLX), or control chow. Treatment was started at 5 weeks of age, before the onset of clinical symptoms and continued up to ∼ 8 months of age (WT, 252 ± 18 and cmo 248 ± 19 days), at which time the mice were euthanized and analyzed. (B-C) Micro CT reconstruction of distal tails (B) and hind paws (C) of PLX-treated and control cmo mice. (D) PLX treatment decreases circulating MIP-1α in cmo mice. (E) Quantitative bone parameters of PLX-treated and control cmo and WT mice. (F) Micro CT reconstructions of proximal tibiae. (G-H) PLX treatment of cmo mice prevents splenomegaly (G) and the expansion of splenic OCP (H). Scale bars, 100 μm. Data ± SEM, n ≥ 3.
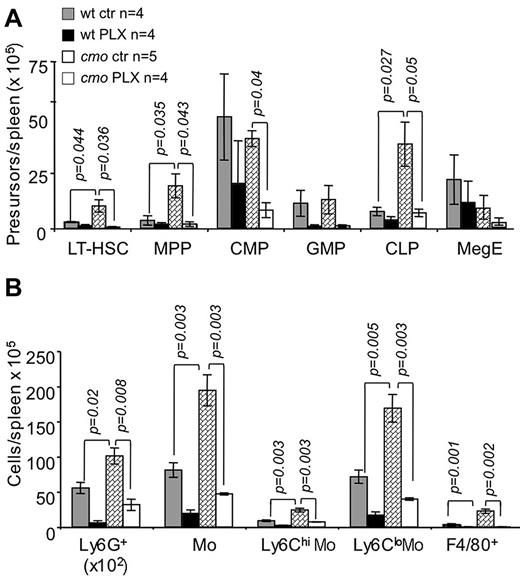
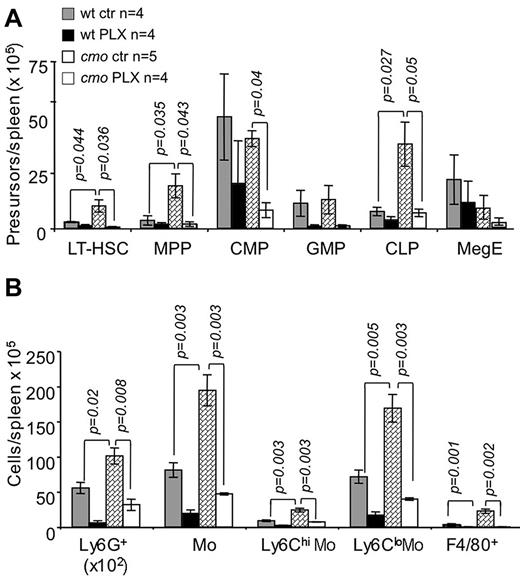
The extramedullary hematopoiesis with splenomegaly (Figure 3G) and the expansion of splenic EOCP, LOCP (Figure 3H), myeloid precursors (Figure 4A), granulocytes, monocytes, and macrophages (Figure 4B) were also inhibited by PLX3397. In contrast, PLX3397 had no effect on early myelopoiesis in bone marrow (Figure 3H and data not shown). These data suggest that increased CSF-1R and c-Kit signaling plays a central role in the development of cmo autoinflammatory disease by promoting myelopoiesis in the spleen.
The dual specificity c-Fms and c-Kit inhibitor, PLX3397, suppresses extramedullary hematopoiesis in cmo mice. PLX3397 prevents the expansion of both early hematopoietic precursors (A) and myeloid cells (B) in spleen. Data ± SEM; n indicates number of mice/group; and ns, not significant (P > .05).
The dual specificity c-Fms and c-Kit inhibitor, PLX3397, suppresses extramedullary hematopoiesis in cmo mice. PLX3397 prevents the expansion of both early hematopoietic precursors (A) and myeloid cells (B) in spleen. Data ± SEM; n indicates number of mice/group; and ns, not significant (P > .05).
PSTPIP2 deficiency leads to increased differentiation of c-Kit+ c-fmslo Mac1lo EOCP
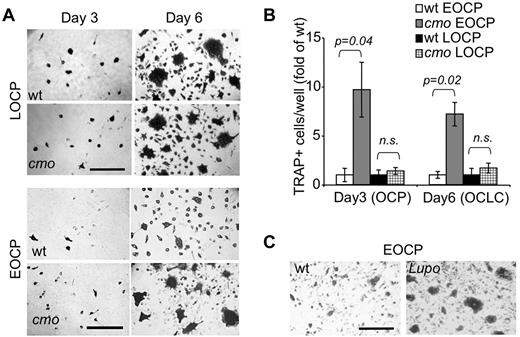
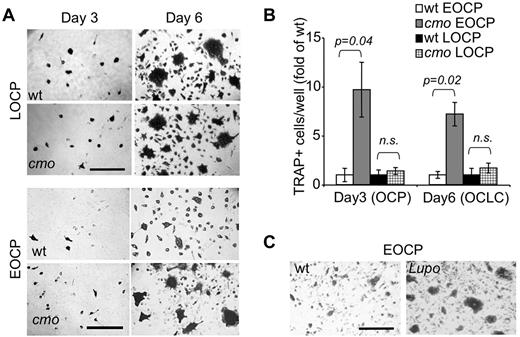
Because the frequencies of bone marrow OC precursors were comparable in WT and cmo mice, yet osteoclasts developed faster in cmo bone marrow cultures than in WT cultures (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), we determined whether PSTPIP2 deficiency enhanced the ability of OC precursors to differentiate by assessing the ability of isolated multipotent c-Kit+ c-Fmslo Mac1lo EOCP and more mature c-Kit+ c-Fmshi Mac1lo LOCP to acquire TRAP expression and to fuse into osteoclasts in vitro, in the presence of CSF-1 and RANKL. The c-Kit+ c-Fmshi Mac1lo LOCP fraction isolated from cmo or WT bone marrow exhibited comparable ability to form osteoclasts (Figure 5A top panels and B). In contrast, the c-Kit+ c-Fmslo Mac1lo EOCP fraction isolated from cmo mice exhibited an increased ability to generate TRAP+ cells and formed larger osteoclasts in vitro compared with the same fraction from WT mice (Figure 5A bottom panels and B). Similar results were obtained using EOCP isolated from Lupo mice (Figure 5C). These data indicate that PSTPIP2 negatively regulates the differentiation of c-Kit+ c-Fmslo Mac1lo EOCP.
PSTPIP2 deficiency enhances osteoclast development from purified precursors. (A) TRAP staining of c-Kit+ c-Fmshi Mac-1lo (LOCP) and c-Kit+ c-Fmslo Mac-1lo (EOCP) osteoclast precursor cultures at day 3 and day 6 of CSF-1 and RANKL-induced osteoclast differentiation in vitro. (B) Quantitation of TRAP+ mononuclear OCP (day 3) or OCLC cells (day 6) shown in panel A. Data ± SEM from 3 independent experiments. ns indicates not significant (P > .05). (C) Compared with WT controls, cultured Lupo EOCP form more TRAP-positive multinucleated osteoclasts.
PSTPIP2 deficiency enhances osteoclast development from purified precursors. (A) TRAP staining of c-Kit+ c-Fmshi Mac-1lo (LOCP) and c-Kit+ c-Fmslo Mac-1lo (EOCP) osteoclast precursor cultures at day 3 and day 6 of CSF-1 and RANKL-induced osteoclast differentiation in vitro. (B) Quantitation of TRAP+ mononuclear OCP (day 3) or OCLC cells (day 6) shown in panel A. Data ± SEM from 3 independent experiments. ns indicates not significant (P > .05). (C) Compared with WT controls, cultured Lupo EOCP form more TRAP-positive multinucleated osteoclasts.
The Lupo mutation (I282N) impairs the function of the PSTPIP2 F-BAR domain and PSTPIP2 stability
The cmo mutation (L98P) is located within the predicted F-BAR domain, which mediates PSTPIP2 interaction with membrane phospholipids and promotes membrane curvature,28 whereas the Lupo mutation (I282N) is located adjacent to the F-BAR domain. In contrast to the cmo mutation which leads to absence of detectable PSTPIP2 protein expression,14 the Lupo mutation leads to partial protein deficiency.11 However, I282 is highly conserved in F-BAR proteins and molecular modeling predicts that substitution of the hydrophobic I282 with a polar residue (Asn) destabilizes the 3D structure by impeding the contact of the unstructured C-terminus with a hydrophobic groove (supplemental Figure 2A and Henne et al29 ). Thus, we investigated the impact of the Lupo mutation on the structure and function of PSTPIP2 in membrane tubulation experiments, which are a standard measure of the membrane-bending activity of BAR and F-BAR domain-containing proteins.29-31 The PSTPIP2 I282N mutation did not significantly affect PSTPIP2 folding in solution (supplemental Figure 2B), nor the ability of PSTPIP2 to tubulate liposomes in vitro (Figure 6A). We therefore examined the formation and stability of tubules formed in cells overexpressing the WT or mutant PSTPIP2, a condition that permits analysis of tubule turnover, as well as interactions of PSTPIP2 with other cellular proteins. PSTPIP2 was followed in these experiments by tagging the N-terminus with GFP, as N-terminal tagging does not affect PSTPIP2 activities in macrophages.24 Membrane tubules induced by the overexpression of the I282N mutant in COS-7 cells were unstable compared with those formed by WT PSTPIP2 (Figure 6B). In general, tubules generated by the I282N mutant were less abundant and typically broke at one end, indicated by a quick loss of fluorescence down the axis of the tubule (Figure 6C). This suggested a fast depolymerization of the tubule PSTPIP2 I282N coat. EGFP-PSTPIP2 I282N (Figure 6D), but not EGFP-PSTPIP2 WT, also appeared to aggregate in transfected cells, forming fluorescent densities throughout the cytoplasm. Aggregates were closely associated with membrane tubules, and some appeared tethered to the tubules in live cell imaging (Figure 6D), suggesting that they are generated at sites of high PSTPIP2 concentration (ie, tubules). These observations indicate that the I282N mutation significantly affects both the ability of PSTPIP2 to stabilize membrane curvature, as well as the stability of PSTPIP2 itself. They also demonstrate that although residual PSTPIP2 is expressed in Lupo mice, it is dysfunctional.
The I282N (Lupo) mutation impairs both membrane sculpting by PSTPIP2 and PSTPIP2 stability. (A) Transmission electron micrographs of Folch liposomes (left panel) and liposomes coincubated with WT PSTPIP2 (middle panel) demonstrate that PSTPIP2 typically generates narrow (∼ 25-nm diameter) tubules. Striations decorate some portions of the tubules (inset), suggesting a PSTPIP2 repeating scaffold. (Right panel) PSTPIP2 l282N mutation does not attenuate tubulation in vitro. (B) Mutation of I282 affects plasma membrane tubule stability. Pseudocolored fluorescence image of COS-7 cells expressing EGFP alone (left), WT EGFP-PSTPIP2 (middle) or EGFP-PSTPIP2 l282N (right) under live cell imaging. The EGFP fluorescence at time = 0 seconds is displayed in green and in the same cell 30 seconds later, in red. The lengths of PSTPIP2 l282N tubules decrease over time (arrows), whereas WT PSTPIP2 tubules appear stable, as indicated by the colocalization of red and green signals (yellow). Note: movement of tubules during the 30 seconds time frame results in separate green and red images for the same tubule. (C) Time-lapse video frames of live cell imaging of a COS-7 cell expressing EGFP-PSTPIP2 I282N at moderate levels. The arrow in the first frame denotes the site of tubule uncoating. (D) At high expression levels EGFP-PSTPIP2 I282N misfolds into large protein aggregates that are tethered to tubules (inset, arrows). Scale bars, 200 nm in panel A and 20 μm in panels B through D.
The I282N (Lupo) mutation impairs both membrane sculpting by PSTPIP2 and PSTPIP2 stability. (A) Transmission electron micrographs of Folch liposomes (left panel) and liposomes coincubated with WT PSTPIP2 (middle panel) demonstrate that PSTPIP2 typically generates narrow (∼ 25-nm diameter) tubules. Striations decorate some portions of the tubules (inset), suggesting a PSTPIP2 repeating scaffold. (Right panel) PSTPIP2 l282N mutation does not attenuate tubulation in vitro. (B) Mutation of I282 affects plasma membrane tubule stability. Pseudocolored fluorescence image of COS-7 cells expressing EGFP alone (left), WT EGFP-PSTPIP2 (middle) or EGFP-PSTPIP2 l282N (right) under live cell imaging. The EGFP fluorescence at time = 0 seconds is displayed in green and in the same cell 30 seconds later, in red. The lengths of PSTPIP2 l282N tubules decrease over time (arrows), whereas WT PSTPIP2 tubules appear stable, as indicated by the colocalization of red and green signals (yellow). Note: movement of tubules during the 30 seconds time frame results in separate green and red images for the same tubule. (C) Time-lapse video frames of live cell imaging of a COS-7 cell expressing EGFP-PSTPIP2 I282N at moderate levels. The arrow in the first frame denotes the site of tubule uncoating. (D) At high expression levels EGFP-PSTPIP2 I282N misfolds into large protein aggregates that are tethered to tubules (inset, arrows). Scale bars, 200 nm in panel A and 20 μm in panels B through D.
Distinct steps of osteoclastogenesis are negatively regulated by PSTPIP2 tyrosine phosphorylation, membrane interactions, or PEST-type phosphatase binding
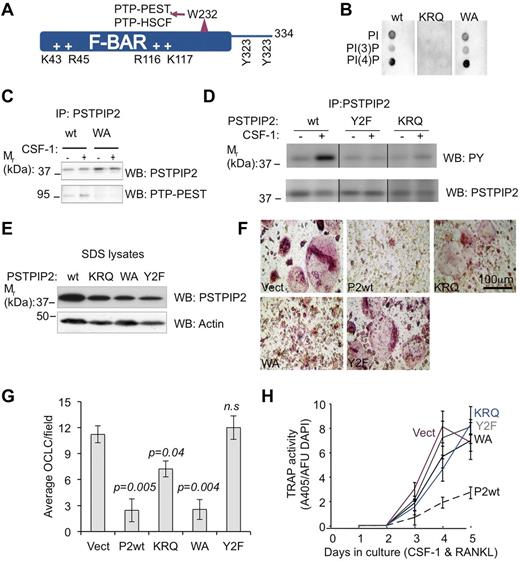
To better understand how PSTPIP2 modulates osteoclast differentiation, we generated an immortalized c-Kit+ c-Fmslo Mac1lo OC precursor cmo cell line (cmoOCP) that failed to express endogenous PSTPIP2 protein. We then reconstituted PSTPIP2 expression in these cells by retroviral transduction of WT PSTPIP2 (P2WT; Figure 7A) and mutants, an R116Q, K117Q mutant unable to interact with membrane phospholipids (KRQ; Figure 7B), a W232A mutant unable to interact with PEST-type phosphatases (WA; Figure 7C), or a mutant that eliminates CSF-1R–stimulated PSTPIP2 tyrosine phosphorylation (Y2F; Figure 7D). The levels of expression of the PSTPIP2 mutants in cells were only slightly lower than the expression of WT PSTPIP2 (Figure 7E), indicating that these mutations did not significantly impair PSTPIP2 stability. After 5 days of culture in CSF-1 and RANKL, cmoOCPs transduced with the vector develop into large multinucleated TRAP+ osteoclast-like cells (OCLC). In contrast, in cultured cmoOCPs expressing WT PSTPIP2, the development of OCLCs was dramatically suppressed (Figure 7F). The WA mutant also suppressed OCLC development, but was less effective at suppressing TRAP expression (Figure 7F-H). In contrast, the Y2F mutant lost the ability to suppress osteoclast development completely (Figure 7F-H). cmoOCPs expressing the KRQ mutant had an intermediate phenotype generating fewer, large, multinucleated TRAP+ cells (Figure 7F-H). Interestingly, compared with WT, the KRQ mutant also exhibited reduced tyrosine phosphorylation but, unlike Y2F, its phosphorylation at CSF-1–regulated sites was increased after stimulation with CSF-1 (Figure 7D right panel). These differences in OC development between cell lines expressing various PSTPIP2 mutants (Figure 7F-H) cannot simply be accounted for by the slight differences in PSTPIP2 expression levels (Figure 7E). For example, the WA mutant was comparable with PSTPIP2 WT in its ability to suppress OCP fusion, whereas the KRQ and the Y2F mutants, expressing PSTPIP2 at levels higher or comparable with those of the WA mutant, were less able to inhibit OCP fusion. We conclude that PSTPIP2 tyrosine phosphorylation and a functional F-BAR domain are essential for its inhibitory actions on TRAP expression and osteoclast precursor fusion, whereas interaction with PEST-type phosphatases (PTP-PEST and PTP-HCSF) is required for suppression of TRAP expression.
PSTPIP2 tyrosine phosphorylation and interaction with membrane phospholipids are required for PSTPIP2 inhibition of osteoclast differentiation. (A) Predicted structural determinants of PSTPIP2 molecular interactions. (B) Mutation of the conserved cationic residues, R116 and K117 (KRQ), abolishes phospholipid binding. (C) Mutation of W232 to alanine (WA) inhibits PSTPIP2 interaction with PTP-PEST. (D) Mutation of the major tyrosine phosphorylation sites Y323 and Y333 (Y2F) eliminates tyrosine phosphorylation of PSTPIP2 in response to CSF-1 stimulation (middle panel), which is also reduced in CSF-1–stimulated macrophages expressing the KRQ mutant (right panel). Repositioned gel lanes from the same blot are separated by vertical lines. (E) The expression of WT PSTPIP2 and PSTPIP2 mutants was confirmed by Western blotting of SDS cell lysates. Actin indicates loading control. (F) Morphology of day 6 osteoclasts obtained from immortalized cmo c-Kit+ Mac-1lo c-Fmslo EOCP retrovirally transduced with vector (Vect), WT PSTPIP2 (P2WT), or the PSTPIP2 mutants described in panels B through E. (G) Quantitation of the number of OCLC per field. (H) Effects of PSTPIP2 deficiency and mutation on TRAP expression. AFU DAPI indicates arbitrary fluorescence units of DAPI-stained cultures. Data ± SEM, n ≥ 3 independent experiments; ns indicates not significant (P > .05).
PSTPIP2 tyrosine phosphorylation and interaction with membrane phospholipids are required for PSTPIP2 inhibition of osteoclast differentiation. (A) Predicted structural determinants of PSTPIP2 molecular interactions. (B) Mutation of the conserved cationic residues, R116 and K117 (KRQ), abolishes phospholipid binding. (C) Mutation of W232 to alanine (WA) inhibits PSTPIP2 interaction with PTP-PEST. (D) Mutation of the major tyrosine phosphorylation sites Y323 and Y333 (Y2F) eliminates tyrosine phosphorylation of PSTPIP2 in response to CSF-1 stimulation (middle panel), which is also reduced in CSF-1–stimulated macrophages expressing the KRQ mutant (right panel). Repositioned gel lanes from the same blot are separated by vertical lines. (E) The expression of WT PSTPIP2 and PSTPIP2 mutants was confirmed by Western blotting of SDS cell lysates. Actin indicates loading control. (F) Morphology of day 6 osteoclasts obtained from immortalized cmo c-Kit+ Mac-1lo c-Fmslo EOCP retrovirally transduced with vector (Vect), WT PSTPIP2 (P2WT), or the PSTPIP2 mutants described in panels B through E. (G) Quantitation of the number of OCLC per field. (H) Effects of PSTPIP2 deficiency and mutation on TRAP expression. AFU DAPI indicates arbitrary fluorescence units of DAPI-stained cultures. Data ± SEM, n ≥ 3 independent experiments; ns indicates not significant (P > .05).
Discussion
The identification of changes in bone density associated with chronic immune activation, including autoimmune and autoinflammatory diseases, cancer, and atherosclerosis, has revealed a dynamic interplay between bone and the immune system and has led to the emergence and rapid development of the new field of osteoimmunology.32,33 In this paper, we focused on the elucidation of the actions of the anti-inflammatory protein PSTPIP2/MAYP11,14 in osteoclastogenesis. We show that the effects of PSTPIP2 deficiency autoinflammatory disease on the skeletal system are not limited to erosive lesions at sites of inflammation, but lead to generalized osteopenia, resulting from increased osteoclast differentiation. These data demonstrate that PSTPIP2 negatively regulates pathways leading to both inflammation and bone resorption.
PSTPIP2 deficiency causes the elevation of the serum levels of the pro-osteoclastogenic factor, MIP-1α. In response to CSF-1, PSTPIP2-deficient macrophages produce more MIP-1α than WT counterparts (Figure 2E, and Chitu et al14 ). This observation, coupled with the increase in macrophage abundance in PSTPIP2-deficient mice (Figure 4B; Grosse et al11 and Chitu et al14 ), suggest that macrophages are the major contributor to the elevation of circulating MIP-1α in the mutant mice. Consistent with this, we found that suppression of myelopoiesis using the dual c-Kit/CSF-1R inhibitor, PLX3397 (Figure 3), prevented the elevation of serum MIP-1α and extramedullary hematopoiesis and attenuated disease. PLX3397 also prevented the trabecular bone loss in cmo disease. However, in contrast to genetic ablation of the CSF-1R19 or antibody-mediated ablation of CSF-134 in developing mice, there was no effect of PLX 3397 on cortical bone thickness, reflecting the slow turnover rate of cortical bone in adult mice.35
MIP-1α is chemotactic for OC precursors in vivo36 and promotes osteoclastogenesis in vitro, via its actions on both CCR1 and CCR5.37 CCR1 is the major MIP-1α receptor expressed in preosteoclasts and osteoclasts and is up-regulated during osteoclast differentiation.36 Studies in CCR1-deficient mice revealed that CCR1 is critical for the mobilization of myeloid precursors and for the enhancement of CSF-1–dependent macrophage colony formation by MIP-1α.38 Furthermore, blocking MIP-1α signaling, using a CCR1 antagonist, inhibits osteoclastogenesis in vitro.39 Together, these data suggest that PSTPIP2 deficiency disease results from enhanced CSF-1 signaling leading to overproduction of monocytic lineage cells that subsequently become activated to produce MIP-1α.
Both cmo (Figure 3G) and 3- to 5-month-old Lupo mice (% splenic weight/body weight 0.48 ± 0.02 compared to WT 0.34 ± 0.02; P = .0003; n = 9 mice/genotype) exhibit splenomegaly. In the case of the cmo mice, the splenomegaly was shown to be due to an overall increase in the numbers of hematopoietic cells, including LT-HSC, MPP, CMP, and CLP (Figure 4A) and their differentiated progeny (Figure 4B). In bone marrow, there was no change except for an increase in Mac1low c-Fmslow c-Kit+ multipotent myeloid precursors. All these cells express c-Fms and/or c-Kit (CLP only express c-Kit). PSTPIP2 is tyrosine phosphorylated downstream of the CSF-1R13,24 and c-Kit,12 and PSTPIP2 expression increases during myeloid differentiation.40 Furthermore, PSTPIP2 inhibits CSF-1–induced macrophage Erk1/2 activation14 and Erk1/2 is a major regulator of CSF-1–induced proliferation in macrophages and their progenitors.41,42 These data suggested that the extramedullary hematopoiesis was because of increased c-Kit and CSF-1R proliferation signaling. Indeed, the expansion of the splenic populations was suppressed by inhibiting c-Kit and c-Fms signaling with PLX3397 (Figures 3H and 4).
The autoinflammatory disease in PSTPIP2-deficient mice resembles human CRMO.15 It is generally agreed that nonsteroidal anti-inflammatory drugs (NSAIDs) constitute the best initial treatment for CRMO, whereas bisphosphonates and TNFα antagonists are effective in the most severe forms.43 However, the treatment has not been standardized. Furthermore, long-term use of NSAIDs is limited by the gastrointestinal side effects, and TNFα antagonists, although effective, increase risk of infection.43 Based on our observations in mice, we suggest that CSF-1R and c-Kit–regulated cells of the monocytic lineage are a therapeutic target for attenuation of inflammation and osteopenia. Although effective in inhibiting the extramedullary hematopoiesis in cmo mice, the treatment with PLX3397 did not significantly impair bone marrow myelopoiesis or decrease the frequency of blood monocytes. These data are consistent with previous observations in CSF-1R and CSF-1–deficient mice, which indicate that the CSF-1R is not essential for many aspects of the immune defense against pathogens, other than at the maternal-fetal interface.4,44 Thus, targeting CSF-1R and c-Kit signaling should be effective in suppressing autoinflammatory disease without impairing immunity.
Bone marrow, spleen, and peripheral blood cell populations can form OCLC in vitro in the presence of CSF-1 and RANKL.45 Stem cell factor-induced up-regulation of the CSF-1R in c-Kit+ c-Fms−/lo Mac-1lo multipotent bone marrow precursors, followed by CSF-1–stimulated RANK expression leads to the differentiation of c-Kit+ c-Fms+ Mac-1lo intermediate osteoclast precursors which then further differentiate to c-Kit− c-Fms+ Mac-1lo RANK+ late osteoclast precursors.8 Our results indicate that in the absence of PSTPIP2, multipotent c-Kit+ c-Fmslo Mac-1lo EOCPs are selectively increased in vivo. Furthermore, when isolated, these cells are more osteoclastogenic than their WT counterparts. Structure-function analysis reveals that PSTPIP2 regulates pathways involved in multiple steps of osteoclast differentiation, via distinct molecular interactions. The Lupo mutation I282N, although not being deleterious for PSTPIP2 expression, impaired the ability of PSTPIP2 to induce stable membrane invagination in COS-7 cells and in vivo leads to a very similar phenotype to the cmo mutation, which abolished PSTPIP2 expression. Although the conformation of PSTPIP2 in solution did not seem to be significantly affected by the I282N mutation, when expressed at high levels in COS-7 cells, PSTPIP2 I282N formed aggregates. This propensity of the Lupo mutant to form inactive aggregates could preferentially affect biologic processes that involve up-regulation of PSTPIP2 expression, such as the late phase of response to LPS in macrophages,11 myeloid differentiation,40,46 and the function of cells that express high levels of PSTPIP2, such as osteoclasts.14
Targeted mutagenesis revealed that although interaction with PEST-type phosphatases was important for inhibition of TRAP expression, both the ability to interact stably with membrane phospholipids and tyrosine phosphorylation, (which presumably creates docking sites for other proteins containing phosphotyrosine-binding domains), were essential for down-regulation of TRAP expression and inhibiting subsequent OCP fusion. Although it remains to be established how membrane modeling by PSTPIP2 contributes to OC development at the molecular level, the fact that both EOCP expressing the KRQ PSTPIP2 mutant, as well as EOCP isolated from mice expressing the F-BAR domain-destabilizing Lupo mutation (I282N), exhibited an increased capacity to form osteoclasts in culture compared with WT counterparts, suggested that stable interaction of PSTPIP2 with the plasma membrane was important for inhibition of OC differentiation. Membrane reshaping is expected to mediate critical events during osteoclast fusion, such as the endocytosis of DC-STAMP, which is a prerequisite for high fusogenic activity,47 and the cellocytosis, that is, cell-cell internalization events leading to plasma membrane fusion and the merging of cytoplasm between the fusion partners.48 PSTPIP2 belongs to the FCH and BAR (F-BAR) family of proteins that have been implicated in clathrin-mediated endocytosis10,29 and cell-to-cell fusion.49 F-BAR proteins promote endocytosis by simultaneously interacting with membrane phospholipids, via their conserved F-BAR domains and with endocytic adaptors and dynamin, via their Src homology 3 (SH3) or muniscin C-terminal μ homology domain (μHD) domains.10,29 PSTPIP2 lacks these domains10 and it is tempting to speculate that it could inhibit the endocytic actions of other F-BAR proteins by competing for phospholipid binding sites without recruiting the other components of the endocytic machinery.
Interestingly, compared with WT PSTPIP2, the KRQ mutant, that had lost the ability to interact with membrane phospholipids, also exhibited a significant reduction in tyrosine phosphorylation. The requirement for membrane targeting of PSTPIP2 for efficient tyrosine phosphorylation is consistent with a previous report indicating that PSTPIP2 is a substrate of Src family kinases,50 and with our observation that CSF-1–induced tyrosine phosphorylation of PSTPIP2 is abolished in macrophages expressing a variant of CSF-1R (CSF-1R Y559F) that is unable to recruit Src family kinases and inhibited by the Src family kinase inhibitor PP2 (data not shown). Although CSF-1 and RANKL are required for osteoclastogenesis,6-9 our results demonstrate that CSF-1R–regulated PSTPIP2 tyrosine phosphorylation is required for suppression of osteoclastogenesis, indicating that PSTPIP2 normally plays a negative feedback role. Further investigations are necessary to determine how CSF-1R triggers the tyrosine phosphorylation of PSTPIP2 and how tyrosine phosphorylation in turn regulates PSTPIP2 activity.
The gene encoding human PSTPIP2 is located on chromosome 18q12 and mutations in this region are associated with autoimmune disorders and psychiatric disease.51-53 Two studies of the PSTPIP2 coding region in small cohorts of patients with CRMO (n = 10) and synovitis, acne, pustulosis, hyperostosis, and osteitis (SAPHO) syndrome (n = 38)54,55 have found rare coding region variants in PSTPIP2 that were not specifically associated with the disease. Given that the penetrance of the cmo and Lupo mutations varies greatly with the genetic background (Grosse et al11 and data not shown), the effect of some of these mutations may have been obscured. Furthermore, as it is decreased PSTPIP2 expression that leads to disease, further investigations with larger cohorts of CRMO patients, focusing on alterations in PSTPIP2 expression are needed.
Our current and earlier11,14 studies describe a mechanism for the development of an autoinflammatory disease affecting skin, joint, and bone, that is triggered by dysregulation of the development of myeloid cells. However, we previously demonstrated the presence of autoantibodies in PSTPIP2 deficiency disease.11 Autoimmunity can be triggered by remnant epitopes generated after degradation of extracellular matrix components by proteases secreted by macrophages.56 Therefore, in a broader context, a mechanism such as we describe here, involving dysregulation of the innate immune system, could contribute to the initiation of autoimmune disease. Clearly, elucidation of the molecular mechanisms underlying regulation by PSTPIP2 are highly relevant to our understanding of how genetic defects affecting early myelopoietic events predispose to chronic inflammation, bone loss and autoimmunity.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Brian West (Plexxikon, Berkeley, CA) for providing expertise on PLX3397 and its effective dosing in mice, Frank Macaluso and Leslie Gunther-Cummins (Analytical Imaging Facility at Einstein) for assistance with the electron microscopic studies, Antonios Aliprantis and the microCT core at Harvard School of Public Health, and Ranu Basu and Robin Sgueglia for technical support.
This work was supported by National Institutes of Health grant RO1 CA26504 (E.R.S.); Albert Einstein College of Medicine Cancer Center grants 5P30-CA13330 (E.R.S.) and K01AR 054486 (V.C.); a New York Community Trust Blood Diseases Grant (V.C.); and an Abbott Rheumatology Scholar Award (J.F.C.). V.N. was supported by a Fulbright award.
National Institutes of Health
Authorship
Contribution: V.C. designed research, performed cytokine, osteoclast formation, and TRAP assays, FACS analysis, electron microscopy, signaling studies, generated the osteoclast cell lines, analyzed the data, and prepared the paper; V.N. performed the implant experiments and histology; J.F.C. performed the CT scan and evaluation; W.M.H. and H.T.M. performed the biochemical characterization of the Lupo mutation; S.N., H.K., and R.H. performed the histology and FACS analysis; M.C.N. provided expertise with CT evaluation and retroviral transduction; and E.R.S. supervised and designed the research, and prepared the paper.
Conflict-of-interest disclosure: E.R.S. has consulted for Plexxikon. The remaining authors declare no competing financial interests.
Correspondence: E. Richard Stanley, Department of Developmental and Molecular Biology, Albert Einstein College of Medicine, 1300 Morris Park Ave, Bronx, NY 10461; e-mail: richard.stanley@einstein.yu.edu; or Violeta Chitu, Department of Developmental and Molecular Biology, Albert Einstein College of Medicine, 1300 Morris Park Ave, Bronx, NY 10461; e-mail: violeta.chitu@einstein.yu.edu.