Abstract
A 2-day method using flow cytometry and FISH for interphase cells was developed to detect monosomy 7 cells in myelodysplastic syndrome patients. The method, Interphase Chromosome Flow-FISH (IC Flow-FISH), involves fixation of leukocytes from blood, membrane permeabilization, hybridization of cellular DNA with peptide nucleic acid probes with cells intact, and analysis by flow cytometry. Hundreds to thousands of monosomy 7 cells were consistently detected from 10-20 mL of blood in patients with monosomy 7. Proportions of monosomy 7 cells detected in IC Flow-FISH were compared with results from conventional cytogenetics; identification of monosomy 7 populations was verified with FACS; and patient and donor cells were mixed to test for sensitivity. IC Flow-FISH allows for detecting monosomy 7 without requiring bone marrow procurement or the necessity of metaphase spreads, and wider applications to other chromosomal abnormalities are in development.
Introduction
Chromosomal aberrations, including balanced translocations, aneuploidy, and major deletions/insertions, arise in a range of hematologic disorders and neoplasms. Conventional cytogenetics and slide-based FISH have provided diagnostic and prognostic tools in the clinical management of myelodysplastic syndrome (MDS),1 chronic lymphoblastic leukemia,2 and acute myeloid leukemia,3 among many other diseases. Karyotype abnormalities may predict response to therapy and consequently are included in most scoring systems for guiding clinical treatment.4,5 For instance, in patients with MDS, the presence of abnormalities in chromosome 7 has been associated with leukemic transformation and adverse prognosis, whereas del(5q) alone and del(20q) alone are among karyotypes predicting better survival.5
Routine clinical diagnosis of aneuploidies and large deletions is performed with G-banding karyotype or, more recently, FISH. G-banding of cells in metaphase permits detection of aneuploidy, translocations, and deletions on all chromosomes simultaneously, but the method is labor-intensive and requires mitotic cells. FISH can be performed more expeditiously on larger numbers of cells6 ; futhermore, as nonmitotic interphase cells are visualized, FISH is useful in chronic lymphoblastic leukemia and related malignant processes with low rates of cell proliferation. FISH has proven more sensitive than G-banding for some chromosomal abnormalities.7-9 FISH limitations include subjective visual analysis of signals in nuclei, the number of cells routinely evaluated (∼ 200 interphase cells), and the requirement to select a panel of probes to test a subset of chromosomal regions.
Early breakthroughs combining flow cytometry with FISH involved isolation of nuclei and hybridization of centromeric chromosome DNA probes (primarily Y chromosome) with DNA of both normal and leukemic cells.10,11 The modern flow-FISH technique of hybridizing sequence-specific peptide nucleic acid (PNA) probes to cellular DNA within cells, and measuring fluorescence by flow cytometry was developed to measure telomere lengths of lymphocyte populations.12 Flow-FISH has since been used to analyze telomere length in early childhood13 and in patients with chronic myeloid leukemia14 and aplastic anemia.15 Extensions of the method have also been developed including Chromosome Flow-FISH, in which chromosomes in suspension are hybridized with centromeric (or telomeric) probes to give chromosome-specific measurements.16
The ability to test for aneuploidy at high sensitivity using flow cytometry and FISH could provide increased value for diagnosis and management of clinical disease. Interphase Chromosome Flow-FISH (IC Flow-FISH) is a method developed to detect chromosome number abnormalities in peripheral blood cells by staining interphase nuclei with a fluorescently labeled chromosome-specific FISH probe after fixing and permeabilizing cells. By directly quantifying fluorescence signals for thousands of peripheral blood cells, the procedure reduces the subjectivity of the existing slide-based interphase FISH assay while dramatically increasing the number of cells examined. Here we describe the method and show results for healthy donors and patients with MDS and monosomy 7. Our results are confirmed with FACS and fluorescence microscopy to capture images of monosomy 7 and normal cells with bound FISH probes.
Methods
Patient and control subjects
Blood samples were collected after written informed consent was obtained in accordance with the Declaration of Helsinki, according to protocols approved by the Institutional Review Board of the National Heart, Lung, and Blood Institute. After preliminary development of the IC Flow-FISH assay with blood from healthy donors, the method was applied to 8 samples from 6 MDS patients with monosomy 7 (age range, 19-79 years). One healthy donor sample was analyzed with each patient sample as a control (total of 8 healthy subjects analyzed).
Conventional cytogenetics
G-banding karyotypes were obtained for all patients. Cells were cultured for 24 or 48 hours in media without stimulation before analysis (Quest Diagnostics).
Blood collection and mononuclear cell isolation
Peripheral blood (10 mL) was collected in a Sodium Heparin Blood Collection Tube (BD Vacutainer), mixed, and diluted with 3 volumes of Dulbecco PBS to 1 volume of blood. In 15-mL conical tubes, 10 mL of diluted blood was layered above 5 mL of Lymphocyte Separation Medium (MP Biomedicals), and tubes were centrifuged for 30 minutes at 550g. Buffy coats were transferred to a 50-mL conical tube and washed twice: in each wash, cells were suspended in 20 mL of DPBS + 1% BSA, centrifuged for 15 minutes at 500g, and resuspended with 1 mL of DPBS + 1% BSA. Cells were filtered if clumps were seen. A cell count was performed. For some samples, cells were frozen in liquid nitrogen for 2-3 months without appreciable degradation.
Fixation and permeabilization
Cells (1 × 107-1.5 × 107) were pipetted in a 15-mL conical tube. For samples in which the transferred volume was > 400 μL, the tube was centrifuged at 300g, and cells were resuspended to a volume of 300-400 μL. Cells were fixed by adding 3 mL of room temperature Carnoy solution (3:1 methanol/acetic acid) with a transfer pipette in a drop-wise manner while vortexing at low speed with each drop. An additional 7 mL of Carnoy fixative was slowly added with a transfer pipette while vortexing intermittently. The sample was incubated in fixative for 60 minutes at room temperature. (Some samples were stored at this point for up to 4 days in Carnoy fixative at −30°C with minor degradation.) Then the sample was centrifuged at 300g for 10 minutes, resuspended in 5 mL of Carnoy fixative, and centrifuged for 5 minutes at 300g. Carnoy solution was aspirated, and the pellet was resuspended in 2 mL of 1× Permeabilization Buffer (9 mL of H20 and 1 mL of 10 × Permeabilization Buffer; eBioscience) and incubated for 30 minutes at 4°C. Next, cells were washed with 10 mL of DPBS + 1% BSA and centrifuged for 12 minutes at 300g. After aspirating the supernatant, the pellet was resuspended in 5 mL of DPBS + 1% BSA and centrifuged for 5 minutes at 300g. The supernatant was aspirated, and cells were resuspended in 500 μL of DPBS + 1% BSA. Some samples were stored for 1-5 days at 4°C at this point with some cell loss but with clear cytometry results.
Hybridization
A cell count was performed with trypan blue. Next, 3 × 105 cells were pipetted in each of 2 microcentrifuge tubes. The volume of cells per tube ranged from 50-150 μL depending on cell loss in previous steps. Cells were centrifuged for 8 minutes at 500g. All but 20-30 μL of the supernatant was removed, and cells were resuspended by vortex. To each of 2 clean microcentrifuge tubes was added 400 μL of hybridization buffer (6.5 mL of 100% formamide, 1 mL of 20 × saline-sodium citrate buffer, 0.5 mL of H20, and 2 mL of 50% dextran sulfate; prepared just before use). In one tube with hybridization buffer was added 2 μL of FITC-conjugated centromeric chromosome 7 probes (Vysis CEP 7 [D7Z1], Abbott). These 2 tubes were placed in a 74°C water bath for 5 minutes to denature the probes. Each hybridization solution with or without probes was pipetted to a tube with cells; the sample without probes was the control. These tubes with cells in hybridization solution were vortexed for 2-3 seconds and placed in a 74°C circulating water bath for 15 minutes, cooled on ice for 1 minute, vortexed gently, and incubated overnight at 42°C on a heating block covered by aluminum foil to prevent exposure to light.
Posthybridization wash
To each sample tube was added 700 μL of 1 × posthybridization wash buffer (12 mL of room temperature formamide, 6 mL of H2O, and 2 mL of 20 × saline-sodium citrate buffer; prepared just before use), followed by incubation for 10 minutes in a 40°C water bath and centrifugation for 10 minutes at 500g. The supernatant was carefully aspirated, leaving ∼ 150 μL of fluid per tube. Cells were resuspended by gentle vortex. Then 700 μL of 1 × post-hybridization wash buffer was again added to each sample, followed by incubation for 10 minutes in a 40°C water bath, centrifugation for 10 minutes at 500g, and aspiration of supernatant, leaving 150 μL of fluid. Cells were resuspended by vortex. Next, 700 μL of DPBS + 1% BSA was added to each sample, followed by incubation for 10 minutes in a 40°C water bath and centrifugation for 10 minutes at 500g. The supernatant was aspirated, leaving 100 μL of fluid, and cells were resuspended. In each tube was added 300 μL of LDS 751 working solution (LDS 751 stock solution: 2 mg of LDS 751, Exciton; 10 mL of methanol. LDS 751 working solution: 121.5 mL of Isoflow Sheath Fluid, Beckman-Coulter; 1.24 mL of 10% BSA; 1.24 mL of RNase T1; 6.1 μL of LDS 751 stock; exposure to light minimized, and stored for 1 month at most). Samples were incubated for at least 1 hour on ice in the dark.
Data acquisition by flow cytometry and cell sorting
A cytometer acquisition signal was triggered on forward scatter at an electronics threshold of 5000. A signal for chromosome 7 probe was measured through a 530/30-nm band pass filter on a log scale. Total DNA content staining of LDS 751 was measured through a 670-nm-long pass filter. The cytometer was calibrated with FITC-conjugated MESF beads. Two drops of beads were mixed with 500 μL of DPBS, and the voltage was adjusted to view all 5 bands. A control sample was used to implement gating. Mononuclear cell populations were captured and gated in a side scatter/forward scatter-area (SSC/FSC-A) plot. In a forward scatter-height/forward scatter-area (FSC-H/FSC-A) scatter plot, single cells were gated apart from clumped cells. Excessive clumps, seen as cells below a diagonal band, indicated an improper Ficoll procedure or improper fixation. A plot of total DNA (as measured by LDS 751) versus SSC was then used to visualize lymphocyte (lower total DNA) and monocyte populations. Next, the signal from probes was viewed on a plot of total DNA/chromosome 7 probe and on a histogram of chromosome 7 probe. The signal from probes was also viewed on a plot of FSC-A/chromosome 7 probe. For each sample, 10 000-15 000 events were recorded from the total DNA/SSC gate. Patient samples were analyzed immediately after control samples. In one experiment, populations of patient cells with lower chromosome 7 probe signal were sorted from cells with higher chromosome 7 probe signal by FACS and the collected cells were centrifuged on slides using the cytospin technique, stained with 4,6-diamidino-2-phenylindole, and viewed by fluorescence microscopy. In another experiment, cells of a patient were mixed with cells of a normal donor in different proportions immediately before data acquisition by flow cytometry.
Data analysis
Flow cytometry data were acquired and analyzed with FloJo flow cytometry software (Version 8.8.7; TreeStar), and further analysis was performed with the R statistical software package using the mixtools library for analyzing finite mixture models.
Results
Flow cytometry data for healthy donors and patients with monosomy 7
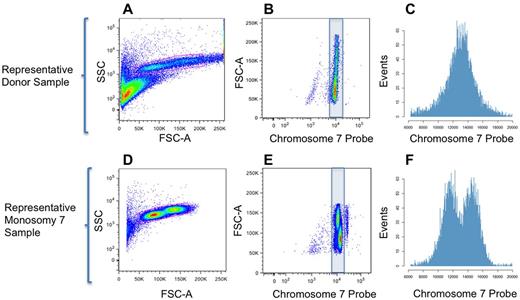
By flow cytometry, 2 populations for each sample corresponding to lymphocytes and monocytes were identified on an SSC versus FSC-A scatter plot (Figure 1A,D). When viewed on a scatter plot of FSC-A versus chromosome 7 probe (or total DNA vs chromosome 7 probe), these populations had similar intensity of probe fluorescence in normal donor samples, with a slight increase in binding as cell size increased (Figure 1B). In samples from MDS patients with monosomy 7, a population of monocytes was observed with lower chromosome 7 probe intensity (Figure 1E-F) than was seen in patient lymphocytes or in normal donor cells (Figure 1B-C). As monosomy 7 occurs in the myeloid rather than the lymphoid lineage in MDS patients,17 our observation of lower probe binding in patient monocytes than in lymphocytes corresponded to expectations for a patient with a high percentage (95%) of monosomy 7 cells by cytogenetics.
Results from IC Flow-FISH. (A,D) Scatter plots of forward and side scatter allowed gating of lymphocytes and monocytes distinct from particulate debris in donor and patient samples. (B,E) Scatter plots of FSC-A versus chromosome 7 probe signal gave vertical separation of (top) monocyte and (bottom) lymphocyte populations based on size. In samples with monosomy 7, some or all monocytes showed lower chromosome 7 probe binding. A small third population, rightmost and unshaded in the monosomy 7 sample plot, was frequently present in patient and donor samples. This population appeared as 2-cell clumps or G2M cells after FACS sorting and fluorescence microscopy (separate sample, K.K., J.W., unpublished data, March 2012). (C,F) Histograms of the raw data corresponding to the blue-shaded regions in panels B,E with a linear axis for probe binding.
Results from IC Flow-FISH. (A,D) Scatter plots of forward and side scatter allowed gating of lymphocytes and monocytes distinct from particulate debris in donor and patient samples. (B,E) Scatter plots of FSC-A versus chromosome 7 probe signal gave vertical separation of (top) monocyte and (bottom) lymphocyte populations based on size. In samples with monosomy 7, some or all monocytes showed lower chromosome 7 probe binding. A small third population, rightmost and unshaded in the monosomy 7 sample plot, was frequently present in patient and donor samples. This population appeared as 2-cell clumps or G2M cells after FACS sorting and fluorescence microscopy (separate sample, K.K., J.W., unpublished data, March 2012). (C,F) Histograms of the raw data corresponding to the blue-shaded regions in panels B,E with a linear axis for probe binding.
The assay was performed for multiple patients; proportions of monosomy 7 monocytes detected are listed in Table 1, which includes fresh blood samples from patients for whom cytogenetic data from a same-day bone marrow sample were available. Monosomy 7 cell percentages from IC Flow-FISH were calculated from total monocytes in each sample as determined by cytometric gating (supplemental Figure 1, see the Supplemental Materials link at the top of the article). Although bone marrow and peripheral blood may not exhibit identical distributions of clonal lines,17 for no patient was the difference in given percentages from IC Flow-FISH and G-banding karyotype statistically significant.
Confirmation of monosomy 7 by FACS and fluorescence microscopy
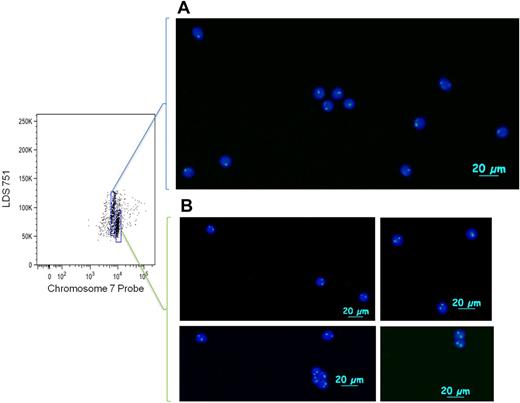
To verify that the population of cells with lower probe fluorescence corresponded to cells with monosomy 7, we sorted post-hybridization cells by FACS (Figure 2). In the lower gate, 2649 cells were collected, and 3556 cells were collected in the higher gate. Viewed by fluorescence microscopy, cells from the gate corresponding to lower probe binding showed 1 distinct binding region in the nucleus in 90% (45 of 50) of cells analyzed and 2 distinct binding regions in 10% (5 of 50) of cells analyzed. Within the gate for higher probe binding, there were 2 distinct nuclear signals in 93% (39 of 42) of cells and 1 distinct nuclear signal in 7% (3 of 42) of cells analyzed.
FACS isolation of monosomy 7 cells after IC Flow-FISH. Monosomy 7 cells (A) and normal cells (B) from PBMCs (frozen for 6 weeks after drawing) were sorted during IC Flow-FISH, stained with 4,6-diamidino-2-phenylindole, and photographed with fluorescence microscopy. Cells were sorted on a BD FACSAria II cell sorter; pictures were taken at room temperature on an Axio Imager D1 (Carl Zeiss Imaging Systems) with 20×/1.4 objective and processed by AxioVision Version 4.6.3.0 (Carl Zeiss Imaging Systems).
FACS isolation of monosomy 7 cells after IC Flow-FISH. Monosomy 7 cells (A) and normal cells (B) from PBMCs (frozen for 6 weeks after drawing) were sorted during IC Flow-FISH, stained with 4,6-diamidino-2-phenylindole, and photographed with fluorescence microscopy. Cells were sorted on a BD FACSAria II cell sorter; pictures were taken at room temperature on an Axio Imager D1 (Carl Zeiss Imaging Systems) with 20×/1.4 objective and processed by AxioVision Version 4.6.3.0 (Carl Zeiss Imaging Systems).
Mixing monosomy 7 patient cells with normal donor cells
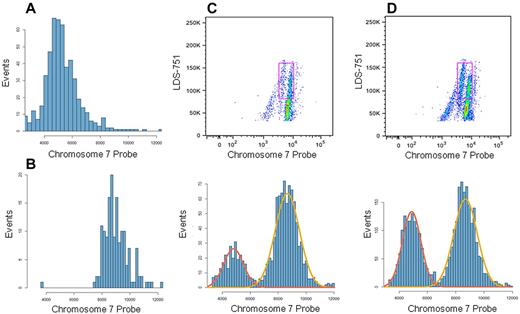
Sensitivity of the assay to differing proportions of monosomy 7 cells was tested by mixing cells of a patient having a high percentage of monosomy 7 (20 of 20 metaphases by cytogenetics) with cells of a normal donor, using target proportions of 5% and 10% patient cells (Figure 3). Actual proportions of patient cells were measured with gating at 6.4% and 14.6% of total cells. After excluding lymphocytes, monosomy 7 was seen in 21% and 38% of total monocytes, as determined by a Gaussian mixture model fit to 1-dimensional data of chromosome 7 probe binding (Figure 3C-D).
Mixing normal donor and monosomy 7 patient cells. Mononuclear cells from an MDS patient with monosomy 7 and from a normal donor were analyzed with IC Flow-FISH independently (without mixing) and after mixing in differing proportions. From flow cytometry data, histograms of chromosome 7 probe fluorescence values were prepared for unmixed patient (A) and unmixed donor (B) monocyte populations. (C-D) Samples having mixtures of patient and donor mononuclear cells, with 5% patient cells expected in panel C and 10% patient cells expected in panel D. Monocytes (cells corresponding to gated regions in scatter plots) were selected, and histograms of monocyte probe fluorescence were prepared as in panels A and B. Proportions of each population were found by applying a 2-component Gaussian mixture model, and the fitted normal curves shown on histograms were generated from the mixture model results. The monosomy 7 populations (lower-binding populations; red normal curve in histograms) were found to represent 21.0% (C) and 38.2% (D) of monocytes.
Mixing normal donor and monosomy 7 patient cells. Mononuclear cells from an MDS patient with monosomy 7 and from a normal donor were analyzed with IC Flow-FISH independently (without mixing) and after mixing in differing proportions. From flow cytometry data, histograms of chromosome 7 probe fluorescence values were prepared for unmixed patient (A) and unmixed donor (B) monocyte populations. (C-D) Samples having mixtures of patient and donor mononuclear cells, with 5% patient cells expected in panel C and 10% patient cells expected in panel D. Monocytes (cells corresponding to gated regions in scatter plots) were selected, and histograms of monocyte probe fluorescence were prepared as in panels A and B. Proportions of each population were found by applying a 2-component Gaussian mixture model, and the fitted normal curves shown on histograms were generated from the mixture model results. The monosomy 7 populations (lower-binding populations; red normal curve in histograms) were found to represent 21.0% (C) and 38.2% (D) of monocytes.
Discussion
IC Flow-FISH offers a means to obtain and analyze higher quantities of data than are commonly gathered in conventional cytogenetics or slide-based FISH, without the need for bone marrow procurement or metaphase spreads. The technique has speed comparable with standard FISH; freshly drawn blood can be processed and data analyzed in 24-30 hours or in stages over several days or weeks. In addition, IC Flow-FISH can facilitate screening tens of thousands of cells per patient, compared with a few hundred cells usually evaluated on a conventional FISH slide. Because quantified fluorescence data can be analyzed by flow cytometric gating and algorithmic mixture modeling, subjective visual examination of individual cells is unnecessary. In practice, IC Flow-FISH could be coupled with other biochemical and molecular analyses, including novel genomic methods, such as comparative genomic hybridization, to identify cell subtypes affected by chromosomal changes as well as the proportions of cells with chromosomal changes. Future applications may include rapid screening of multiple probes in one sample and downstream characterization of sorted cells with aneuploidy, including sequence analysis or array comparative genomic hybridization to map patterns of neoplastic clonal evolution among populations of cells with chromosome number abnormalities.
Here the method has been clinically applied to patient blood samples for detection and isolation of monosomy 7 cells. Proportions of monosomy 7 cells detected by IC Flow-FISH were consistent with results from conventional cytogenetics for all patient samples, and the use of FACS and microscopy confirmed the subpopulations of monosomy 7 and normal cells. When applied to mixed samples, the method allowed for clear distinction between the differing proportions of monosomy 7 cells, as assessed by algorithmic modeling.
The Vysis CEP 7 (D7Z1) FITC-conjugated SpectrumGreen probe gave reliable DNA staining with low signal variability among samples. Initially, the procedure was developed using centromeric probes for the X and Y chromosomes from healthy persons (supplemental Figure 2). Extension to other chromosomal abnormalities is in progress, with promising results for 13q− in chronic lymphoblastic leukemia patient samples (K.K., J.W., R.T.C., unpublished data, January 2012). Identification of viable commercial PNA probes specific to various centromeric and noncentromeric chromosomal regions is an area of active development. Extension to other tissues may be complicated by multiple factors. As observed by Wieser et al examining kidney cells, the presence of autofluorescence in cell cytoplasm can affect recorded fluorescence during data acquisition in flow cytometry.18 In all cases, plasma and nuclear membrane penetration, as well as heat stability of the probe, are primary considerations.
Whereas best-preserved cell morphologies were seen with fresh samples, Ficolled samples frozen in liquid nitrogen for 1-2 months showed little to no signs of degradation. Cells stored for over a year degraded under the conditions of fixation and heating. Permeabilized and washed cells could be stored in DPBS + 1% BSA at 4°C for up to 5 days before hybridization with minimal degradation. Fixed cells could also be stored in Carnoy fixative at −30°C for 3-4 days before proceeding with the assay, although minor degradation was apparent. Clearest results were obtained when careful attention was paid to cell handling, including maximal removal of red blood cells from the sample to minimize cellular debris, which has the potential to nonspecifically bind PNA probes. In samples with clumping of leukocytes before fixation, significant cell loss was seen.
Combining FISH staining with antibody phenotyping has the potential for more clearly identifying leukocyte subpopulations. However, preliminary results show that this approach was complicated by loss of antibody fluorescence during fixation, and paraformaldehyde fixation of antibodies gave low and heterogeneous PNA probe binding after hybridization (K.K., J.W., unpublished data, November 2011). Replacing conventional fluorophores with quantum dots has been shown to better preserve fluorescence in conditions of high heat for flow-FISH19 and may be optimal.
The appearance of monosomy 7 and other cytogenetic abnormalities in hematologic disorders can substantially change patient prognosis and treatment considerations. As shown here, IC Flow-FISH is a technique to gather data on chromosome number abnormalities for hundreds to tens of thousands of cells from peripheral blood. The method has been effective and consistent in detecting monosomy 7 in MDS patients; application to other chromosome number abnormalities is currently in development.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Bogdan Dumitriu and Ankur Parikh (National Heart, Lung, and Blood Institute, National Institutes of Health) for reviewing the manuscript and DeLong Liu for statistical advice (Mathematical and Statistical Computing Laboratory, CIT, and Genomics Core Facility, National Heart, Lung, and Blood Institute, National Institutes of Health).
This work was supported by the National Institutes of Health Intramural Research Program. R.T.C. was supported in part by Fundacao de Amparo a Pesquisa do Estado de São Paulo.
National Institutes of Health
Authorship
Contribution: K.K. developed the method, designed and performed experiments, and wrote the manuscript; J.W. designed and performed experiments, analyzed data and programmed, and wrote the manuscript; P.S. designed and performed experiments; S.K. contributed to writing the manuscript; R.T.C. conceived the method and contributed to writing the manuscript; and N.S.Y. conceived the method and experiments, analyzed results, and contributed to writing the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jason Weed, Hematology Branch, National Heart, Lung, and Blood Institute, National Institutes of Health, Bldg 10-CRC, Rm 3E-5216, 9000 Rockville Pike, Bethesda, MD 20892; e-mail: jason.weed@nih.gov.
References
Author notes
K.K. and J.W. contributed equally to this study.