Abstract
Recent studies have established that during embryonic development, hematopoietic progenitors and stem cells are generated from hemogenic endothelium precursors through a process termed endothelial to hematopoietic transition (EHT). The transcription factor RUNX1 is essential for this process, but its main downstream effectors remain largely unknown. Here, we report the identification of Gfi1 and Gfi1b as direct targets of RUNX1 and critical regulators of EHT. GFI1 and GFI1B are able to trigger, in the absence of RUNX1, the down-regulation of endothelial markers and the formation of round cells, a morphologic change characteristic of EHT. Conversely, blood progenitors in Gfi1- and Gfi1b-deficient embryos maintain the expression of endothelial genes. Moreover, those cells are not released from the yolk sac and disseminated into embryonic tissues. Taken together, our findings demonstrate a critical and specific role of the GFI1 transcription factors in the first steps of the process leading to the generation of hematopoietic progenitors from hemogenic endothelium.
Introduction
Hematopoietic stem cells (HSCs) are pivotal to the continuous generation and maintenance of mature blood cells throughout adult life. Although they reside mainly in the bone marrow at the time of birth and thereafter, they are initially generated during embryonic life in large blood vessels located in the developing embryo.1 Recent studies in mice2-4 and zebrafish5,6 have established that hematopoietic progenitors and stem cells are produced at these sites from endothelial cells by a process termed endothelial to hematopoietic transition (EHT).
Besides these in vivo studies, the in vitro differentiation of embryonic stem (ES) cells represents a powerful system to study these early events of hematopoietic development. Using this system, it was shown that blood cell development is initiated by a clonal hemangioblast precursor, which gives rise to blast colonies with both endothelial and hematopoietic cells,7 a finding subsequently confirmed in vivo.8 More recently, hemangioblasts were shown to generate first a hemogenic endothelium intermediate cell population, which subsequently gives rise to hematopoietic progenitors.9 During this process, hemogenic endothelial cells lose their endothelial identity by altering their flat, adherent appearance into the characteristic round shape of mobile hematopoietic precursor cells.9,10 This transition critically relies on the presence of the transcription factor RUNX1.3,9 Although these studies have together clearly established that the EHT process is the pivotal event in the generation of the first blood cells,9,10 the molecular and cellular mechanisms orchestrating this critical transition remain essentially unknown.
To identify downstream RUNX1 effectors that drive the development of hematopoietic precursor cells from the hemogenic endothelium, we compared the transcriptomes of Runx1+/− and Runx1−/− hemogenic endothelium and identified the Gfi1 and Gfi1b genes as direct targets of RUNX1. Gfi1 and Gfi1b genes encode 2 highly homologous nuclear zinc finger proteins that function as transcriptional repressors.11,12 A C-terminal domain containing 6 C2-H2–type zing finger motifs mediates the DNA-binding activity of these proteins while their repression activity requires the N-terminal SNAIL/GFI-1 (SNAG) domain.12,13 Gfi1 is expressed in many hematopoietic cell types including HSCs, lymphoid and myeloid progenitors as well as mature lymphoid and myeloid cells.14-16 GFI1 deficiency leads mainly to neutropenia17 and reduction in HSC self-renewal capacity.18,19 On the other hand, its paralogue Gfi1b is mostly expressed in erythroid and megakaryocytic lineages,20 and Gfi1b knockout results in embryonic lethality at embryonic day 14.5 (E14.5) because of a deficiency in erythroid and megakaryocyte development.21 Several studies have shown that the expression of Gfi1 and Gfi1b is controlled by negative feedback loops and that they can also repress expression of each other in a cross-regulatory fashion.16,22-24
We demonstrate here that in the absence of RUNX1, GFI1 and GFI1B are able to trigger the loss of the endothelial identity of hemogenic endothelium. Upon Gfi1 or Gfi1b expression, these cells down-regulate the expression of endothelial genes and undergo the morphologic changes observed in the transition from endothelium adherent cells into hematopoietic round cells. However, these cells are unable to acquire the full competence to generate hematopoietic colonies. Conversely, we establish that blood progenitors generated in Gfi1 and Gfi1b double knockout embryos maintain the expression of endothelial genes and cannot be released from their cell layer within the yolk sac, and therefore fail to disseminate into embryonic tissues. Taken together, our findings identify a new and unexpected role of GFI1 and GFI1B transcription factors in mediating the loss of endothelial identity as the first step in the generation of hematopoietic progenitors from the hemogenic endothelium.
Methods
ES cell growth and differentiation
Embryo generation
Timed mating of Gfi1+/GFPGfi1b+/GFP mice was set up and the morning of vaginal plug detection was considered day 0.5. Gastrulating embryos were staged by morphologic landmarks. Genotyping was performed as published previously.16,20 All animal work was performed under regulation in accordance with the United Kingdom Animal Scientific Procedures Act (ASPA) 1986.
BL-CFC and hemogenic endothelium cultures
Flk-1+ EB cells were plated in methylcellulose, on Matrigel (BD Biosciences) or gelatin, in the blast colony-forming cell (BL-CFC) media described previously.27 For hemogenic endothelium culture, TIE2+c-KIT+CD41− cells were cultured on gelatin (cells isolated from day 2 or day 3 BL-CFC cultures). Cultures were maintained in a humidified chamber in a normal O2 (5%) 5% CO2-air mixture at 37°C.
Chromatin immunoprecipitation
After 24 hours of hemogenic endothelium culture in the presence of doxycycline (dox), iRunx1 endothelial cells were cross-linked for 10 minutes at room temperature with 1% formaldehyde (Pierce) and quenched with a one-tenth volume of 2M glycine. Nuclei were prepared as described before,28 sonicated using a Bioruptor water bath in immunoprecipitation buffer I (25mM Tris 1M, pH 8, 150mM NaCl, 2mM EDTA, pH 8, 1% Triton X-100, and 0.25% SDS). After centrifugation, the sheared 0.5- to 2-kb chromatin fragments (1-2 × 106 cells) were diluted with 2 volumes of immunoprecipitation buffer II (25mM Tris, pH 8, 150mM NaCl, 2mM EDTA, pH 8, 1% Triton X-100, 7.5% glycerol), and precipitation was carried out for 2 hours at 4°C using 2 μg of anti-HA Ab (H6908; Sigma-Aldrich) coupled to 15-μL Protein G dynabeads. Beads were washed with low-salt buffer (20mM Tris 1M, pH 8, 150mM NaCl, 2mM EDTA, pH 8, 1% Triton X-100, 0.1% SDS), high-salt buffer (20mM Tris, pH 8, 500mM NaCl, 2mM EDTA, pH 8, 1% Triton X-100, 0.1% SDS), LiCl buffer (10mM Tris, pH 8, 250mM lithium chloride, 1mM EDTA, pH 8, 0.5% NP40, 0.5% sodium deoxycholate) and Tris-EDTA pH 8 containing 50mM sodium chloride. The immune complexes were eluted in 100 μL of elution buffer (100mM NaHCO3, 1% SDS) and, after adding 5 μL of 5M NaCl and proteinase K, the cross-link was reversed at 65°C overnight. DNA was extracted by using the Ampure PCR purification kit and analyzed by quantitative PCR. Signals obtained were divided through the input control and normalized to an internal control region on chromosome 2. Primers are listed in supplemental Table 2 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Flow cytometry and cell sorting
Staining was done as described previously29 and analyses were performed with a FACSCalibur or LSRII (BD Biosciences). Sorts were performed with a FACSAria or a BD Influx (BD Biosciences) or by using magnetic sorting (Miltenyi Biotec). mAbs and streptavidin used were FLK-1-bio, TIE2-bio (TEK4; eBioscience), TIE2-PE, c-KIT-APC, and c-KIT-APC-eFluor780 (2B8; eBioscience), CD41-PE (MWReg30; BD Biosciences), CDH5–Alexa Fluor 647 (eBioBV13; eBioscience) and Strep-PECy7 (eBioscience). FACS data were analyzed using FlowJo software (TreeStar).
Retroviral transduction experiments
VSVg-pseudotyped MSCV retroviruses were produced by using a self-inactivating vector system.30 In brief, viral supernatant was harvested from HEK293T cells at days 2 and 3 after transfection of 3 plasmids with Novagen GeneJuice: pHIT60 (gag/pol), pRV67 (VSVg protein Env), and the MSCV vector expressing the gene of interest (see Figure 1), passed through a 0.45-μm filter (Millipore), concentrated by ultracentrifugation (2 hours, 20 000g, 4°C), and resuspended in PBS. Sorted Runx1−/−Flk1+ EB cells were transduced at a multiplicity of infection of 25 in BL-CFC media and plated in 24-well plates coated with Matrigel at a density of 104 cells/well. After 4 days, the number of colonies of round cells was counted, and cells were isolated for gene expression analysis.
Gene expression analysis
Gene-specific PCR was performed as described previously.9 Primers are listed in supplemental Table 3. Quantitative PCR was performed either with the Light Cycler 480 (Roche) using the Roche SYBR-Green master mix (primers listed in supplemental Table 4) or with the ABI 7900 system (Applied Biosystems) using the universal probes library from Roche (primers and probes listed in supplemental Table 5).
Immunofluorescence
Hemogenic endothelium culture was performed in ibidi μ-slide VI0.4 for 3 days in the presence or absence of dox. Staining was performed as described previously.9 Briefly, uncoupled rat anti–mouse CDH5 (eBioBV13; eBioscience), goat anti–rat-Alexa555 (Invitrogen) and rat anti–mouse CD41-APC (MWReg30; eBioscience) Abs were successively incubated for 1 hour with the fixed cells. The ibidi slide was mounted with ProLong Gold antifade reagent with DAPI (4′,6-diamidino-2-phenylindole; Invitrogen), analyzed using a Zeiss Axiovert 200M microscope, and processed with Metamorph software (Universal Imaging). Overlay of the different channels was performed using the ImageJ software.
Immunohistochemistry
Four-millimeter-thick, formalin-fixed, paraffin-embedded, yolk-sac sections were dewaxed in xylene and rehydrated through graded alcohol. After epitope retrieval in DAKO target retrieval solution (98°C 15 minutes, followed by 15 minutes cooling) and endogenous peroxidase blocking in 0.3% H2O2 for 10 minutes, sections were then blocked in 5% normal goat serum for 20 minutes before the addition of the GFP (Santa Cruz Biotechnology) Ab (1 mg/mL) for 1 hour at room temperature. Mouse IgG2a was used as an isotype control at the same concentration. After 2 × 5-minute washes in TBS, DAKO Envision+ was applied to the section for 30 minutes at room temperature. After another 2 × 5-minute TBS wash, DAB (diaminobenzidine; DAKO) was added for 5 minutes. After a water wash, sections were counterstained in Gills hematoxylin for 1 minute before dehydration, clearing, and coverslipping.
Apoptosis test
The apoptosis test was performed according to the instructions of the manufacturer (annexin V staining protocol, BD Bioscience). Briefly, cells were harvested and stained with anti annexin V–APC (BD Bioscience), anti–mouse CD41-PE (MWReg30; BD Bioscience) and the nucleic acid dye Sytox Blue (Invitrogen). The staining was then analyzed with a FACSAria (BD Bioscience).
Whole mount staining and Heartbeat movies
Whole-mount immunofluorescence staining was performed as previously described.31 E10.5 embryos were stained with biotinylated rat anti–mouse CD31 (BD Biosciences) and rabbit anti-GFP (MBL) Abs used with Alexa Fluor 555–conjugated streptavidin (Invitrogen) and Alexa Fluor 647 goat anti–rabbit IgG (H+L; Invitrogen), respectively. Specimens were analyzed with Leica TCS LSI Macro Confocal, and videos were generated using LAS AF Lite software and processed using Quicktime Pro. For whole-mount immunohistochemical staining, CD31 was counterstained with Avidin HRP (Invitrogen) and was analyzed using a Leica MZ FLIII microscope. Heartbeat videos were taken using Leica DM13000 B and LAS AF Lite software and were processed using Quicktime Pro.
Time-lapse photography
Performed as described previously.9 Phase-contrast images were taken every 3 minutes (supplemental Videos 1 and 2), 2 days after the beginning of the culture, for 24 hours with the image capture and processing software MetaMorph (Universal Imaging). Video animations were made using the software Imaris (Bitplane).
Results
To identify downstream effector genes of RUNX1 implicated in the regulation of the EHT, Runx1+/− and Runx1−/− ES cells were differentiated in vitro to produce hemogenic endothelial cells. Runx1−/− ES cells are able to differentiate into the hemogenic endothelium, but these cells are unable to undergo the morphologic changes characteristic for the EHT. The transcriptomes of these 2 subpopulations were then analyzed using cDNA expression microarrays as previously published.32 We screened the results for differentially expressed genes encoding either transcriptional activators or repressors, which could act downstream of RUNX1 to trigger a switch between endothelial and hematopoietic cell fates. We found that the genes encoding Gfi1 and Gfi1b transcriptional repressors displayed significantly lower expression levels in the absence of RUNX1 (supplemental Figure 1). Although previous studies have clearly established some critical functions for GFI1 and GFI1B at later stages of hematopoiesis,17,21 their expression at the onset of hematopoietic development suggested another as-yet unexplored function during blood specification.
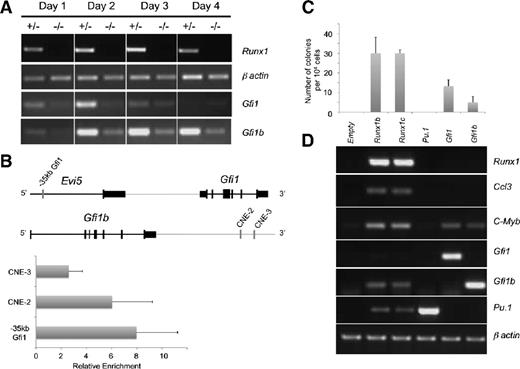
To confirm the microarray results, Gfi1 and Gfi1b expression patterns were determined over a 4-day time course of blast colony development from Runx1+/− or Runx1−/− cells (Figure 1A). In Runx1+/− cells, the expression of Gfi1 and Gfi1b increased progressively during the first 2 days, corresponding to the establishment of the hemogenic endothelium. During the following days of hematopoietic development, the expression of Gfi1b was maintained at a high level while Gfi1 expression strongly decreased. In the absence of RUNX1, expression levels of both genes were dramatically reduced at all time points. Interestingly, we have recently shown that, in hematopoietic progenitors, RUNX1 binds to Gfi1 and Gfi1b regulatory elements (supplemental Figure 2).33,34 By chromatin immunoprecipitation, we additionally demonstrated here binding of RUNX1 to similar elements in hemogenic endothelial cells (Figure 1B). Taken together, these results indicate that RUNX1 directly regulates the expression of Gfi1 and Gfi1b at the onset of hematopoietic development.
Gfi1 and Gfi1b are RUNX1 transcriptional targets in the early stages of blood development. (A) Semiquantitative RT-PCR analysis of Runx1, Gfi1, Gfi1b expression during blast colony development with Runx1+/− and Runx1−/− cells. β actin is the loading control. (B) Chromatin immunoprecipitation analysis showing the direct binding of RUNX1 on Gfi1 and Gfi1b enhancers. (C-D) Day 3.5 Runx1−/− FLK1+ cells were transduced with the indicated retroviruses. Four days after culture, (C) the number of colonies was counted and (D) semiquantitative RT-PCR analysis of expression of the indicated genes was performed. β actin is the loading control. Values in the histogram correspond to an average of 3 transductions for each construct; the error bars correspond to the standard deviation.
Gfi1 and Gfi1b are RUNX1 transcriptional targets in the early stages of blood development. (A) Semiquantitative RT-PCR analysis of Runx1, Gfi1, Gfi1b expression during blast colony development with Runx1+/− and Runx1−/− cells. β actin is the loading control. (B) Chromatin immunoprecipitation analysis showing the direct binding of RUNX1 on Gfi1 and Gfi1b enhancers. (C-D) Day 3.5 Runx1−/− FLK1+ cells were transduced with the indicated retroviruses. Four days after culture, (C) the number of colonies was counted and (D) semiquantitative RT-PCR analysis of expression of the indicated genes was performed. β actin is the loading control. Values in the histogram correspond to an average of 3 transductions for each construct; the error bars correspond to the standard deviation.
To investigate the potential functions of Gfi1 and Gfi1b in the EHT, we evaluated their ability to rescue defects observed in Runx1−/− differentiation cultures. We previously established that Runx1−/− FLK1+ hemangioblast precursors generate fewer blast colonies consisting of round cells.9,27 Runx1−/− FLK1+ cells were transduced with retroviral vectors expressing either Sfpi1 (Pu.1), Runx1, Gfi1, or Gfi1b and assessed for their potential to support the formation of colonies of nonadherent individualized cells. Strikingly, both Gfi1 and Gfi1b were able to support the formation of colonies with such morphology (supplemental Figure 3, Figure 1C), albeit in smaller numbers than with retroviruses expressing either Runx1b or Runx1c, the proximal and distal isoforms of Runx1.35 In contrast, Pu.1 (Sfpi1), a known target of RUNX1 in early hematopoiesis,36 was unable to rescue blast colony formation. Infection with Runx1-expressing viruses rescued the expression of Pu.1, Gfi1b, Ccl3,37 and c-Myb38 , all of which are hematopoietic genes downstream of RUNX1 (Figure 1D). In contrast, rescue with Gfi1- or Gfi1b-expressing vectors only induced the expression of c-Myb (Figure 1D), but not the expression of the other tested hematopoietic genes. The absence of Gfi1 expression after 4 days of Runx1 rescue was consistent with its normal pattern of expression that peaks at day 1 and decreases thereafter (Figure 1A). Overall, these results suggest that both Gfi1 and Gfi1b can support the development of colonies with a typical blast colony morphology.
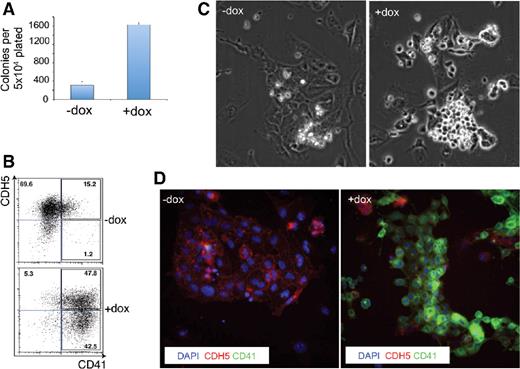
We next investigated the specific steps reinstated by Gfi1 in the absence of Runx1 in more detail. In vitro, the EHT is initiated by RUNX1 in a subpopulation of hemogenic endothelial cells defined by the coexpression of the receptor tyrosine kinase c-Kit (KIT), and the endothelial markers Tek (TIE2) and VE-cadherin (CDH5).9 Initially, these cells do not express the early hematopoietic marker CD41 (also known as Itga2b) and the EHT process begins with a gain of CD41 expression, a stage at which the cells are still adherent.9,10,39 This is then followed by a loss of endothelial markers that leads to the alteration of the endothelial cellular morphology into a round, nonadherent shape. We therefore assessed the ability of Gfi1 to rescue each of these developmental steps using a Runx1−/− ES cell line in which Gfi1 expression, linked to an IRES-GFP, is efficiently induced by doxycycline (referred to as iGfi1 Runx1−/− thereafter). Culture of iGfi1 Runx1−/− FLK1+ cells in the presence of doxycycline resulted in an increased number of colonies with round individualised cells (Figure 2A). This was accompanied by an enhanced frequency of CD41+ cells and a decrease in the frequency of cells coexpressing the endothelial markers TIE2 and CDH5 (data not shown). To determine whether the rescue took place in hemogenic endothelium, a stage at which hematopoietic development is halted in the absence of Runx1, we purified hemogenic endothelium precursors by cell sorting and replated them in the presence or absence of dox. After 3 days of culture, the majority of induced cells were positive for CD41 expression and nearly half of them lacked the expression of the endothelial marker CDH5 (Figure 2B). This was accompanied by changes in cell morphology with the emergence of round, nonadherent cells negative for CDH5 and expressing CD41 (Figure 2C-D, supplemental Videos 1 and 2). However, when we tested the ability of these round cells obtained after Gfi1 induction to generate hematopoietic progeny, we found that they were unable to form any hematopoietic colonies in clonogenic assays (data not shown).
Gfi1 partially rescues Runx1−/− deficiency. (A) Number of colonies generated in methylcellulose by iGfi1 Runx1−/− FLK1+ cells either in absence or presence of dox. (B) FACS analysis of CD41/CDH5 of iGfi1 Runx1−/− TIE2hiKIT+CD41− endothelial cells after 3 days of culture with or without dox. Numbers indicate percentages. (C) Bright field images of endothelial culture after 3 days with or without dox. Pictures were taken with a 10× objective. (D) Immunofluorescence analysis of CD41/CDH5 expression of iGfi1 Runx1−/− hemogenic endothelial cells after 3 days of culture in absence (left panel) or presence (right panel) of dox. Blue corresponds to nuclear (DAPI), red to CDH5, and green to CD41 staining, respectively. These images are overlays of DAPI, CDH5, and CD41 staining. Pictures were taken with a 40× objective.
Gfi1 partially rescues Runx1−/− deficiency. (A) Number of colonies generated in methylcellulose by iGfi1 Runx1−/− FLK1+ cells either in absence or presence of dox. (B) FACS analysis of CD41/CDH5 of iGfi1 Runx1−/− TIE2hiKIT+CD41− endothelial cells after 3 days of culture with or without dox. Numbers indicate percentages. (C) Bright field images of endothelial culture after 3 days with or without dox. Pictures were taken with a 10× objective. (D) Immunofluorescence analysis of CD41/CDH5 expression of iGfi1 Runx1−/− hemogenic endothelial cells after 3 days of culture in absence (left panel) or presence (right panel) of dox. Blue corresponds to nuclear (DAPI), red to CDH5, and green to CD41 staining, respectively. These images are overlays of DAPI, CDH5, and CD41 staining. Pictures were taken with a 40× objective.
To further evaluate the extent of the rescue by GFI1, we examined the expression of endothelial and hematopoietic genes in the populations generated on culture of iGfi1 Runx1−/− hemogenic endothelium cells in the presence or absence of doxycycline (Figure 2B). The CDH5+CD41− (no dox) and CDH5−CD41+ (generated in presence of dox) cell populations were isolated. In addition, CDH5−CD41+ cells generated from normal ES cells after 2 days of hemangioblast culture were used as a control. In agreement with the shutdown of CDH5 expression observed by FACS, endothelial genes such as Cldn5, Pecam1, Eng (endoglin) were down-regulated on Gfi1 expression (supplemental Figure 4). In contrast, expression of other endothelial genes such as Cd34, Kdr (Flk-1), and Tie2 remained elevated. We also investigated expression in these early cell populations of hematopoietic genes such as Gata1, Tal, cMyb, and Lmo2 that are normally expressed at the onset of hematopoietic commitment. We found that they were significantly expressed in the round cells generated on Gfi1 expression. Furthermore, we detected the βH1 hemoglobin transcript indicating the presence of primitive erythroid cells in this population. These results suggest that Gfi1 expression in Runx1−/− hemogenic endothelium cells led to the down-regulation of the expression of some endothelial markers and the concomitant up-regulation of early hematopoietic markers. However, the round cells generated are unable to form any hematopoietic colonies in clonogenic assays. In the absence of RUNX1, a few rare cells emerge from the hemogenic endothelium cell population but they quickly die.6,9 We therefore considered the possibility that the CD41+ cells generated on Gfi1 expression were particularly prone to apoptosis, potentially explaining their inability to produce any hematopoietic colonies. However, when we tested this hypothesis, we did not observe any abnormal rate of apoptosis in these cells (supplemental Figure 5). Finally, to determine whether these changes were correlated with the repressive activity of Gfi1, we generated a Runx1−/− ES cell line with an inducible variant of Gfi1 carrying a mutation in the SNAG domain (P2A) known to completely abolish its repressor activity13 (referred to as iP2AGfi1 Runx1−/− thereafter). This mutated Gfi1 was unable to induce any change in morphology or in hematopoietic or endothelial marker expression (supplemental Figure 6), suggesting that GFI1 repressive activity is critical in this process.
The findings that the GFI1 and GFI1B transcription factors act downstream of RUNX1 in the EHT process suggest that Gfi1 or Gfi1b deletion should recapitulate key aspects of the phenotype observed in Runx1-deficient embryos that die by midgestation (E12.5-13.5) and are devoid of definitive hematopoietic precursors.25,40 However, deletion of either gene individually does not phenocopy Runx1 deficiency.17,21 The lack of an earlier phenotype might be because of coexpression of both proteins in the same cells, allowing a functional compensation for the loss of 1 gene. Indeed, Gfi1 and Gfi1b genes are functionally interchangeable as Gfi1b can rescue the phenotype of the Gfi1 knockout in hematopoiesis.41 Gfi1 and Gfi1b are normally expressed in different subsets of hematopoietic cells with the exception of coexpression in some lymphoid cells20 and HSCs.14 To investigate the potential presence of cells coexpressing both Gfi1 and Gfi1b at the onset of hematopoiesis, we made use of 2 reporter mouse lines with GFP expressed under the control of Gfi1 or Gfi1b promoter, respectively.16,20 We examined by flow cytometry the expression patterns of Gfi1 and Gfi1b in the context of CD41, KIT, TIE2, and FLK1 expression, all indicative of EHT. In E8.5 Gfi1+/GFP embryos, very few GFP+ cells were detected but they all had a Tie2hiKit+FLK1+CD41+ phenotype (Figure 3A middle panels). At the same developmental stage, in Gfi1b+/GFP embryos, a higher frequency of GFP+ cells was detected and these cells were localized in the ring formed by the blood islands in the yolk sac (Figure 3). Most of CD41+ cells were GFP+ including the Tie2hiKit+FLK1+CD41+ subpopulation (Figure 3A bottom panels). Together these findings suggest that both genes are expressed in the Tie2hiKit+FLK1+CD41+ cell population of E8.5 embryos, corresponding to cells becoming hematopoietic progenitors.
Gfi1 and Gfi1b are expressed at the onset of hematopoiesis in vivo. (A) FACS analysis of CD41+GFP− (wild type; top panels), CD41+Gfi1-GFP+ (Gfi1+/GFP; middle panels), and CD41+Gfi1b-GFP+ (Gfi1b+/GFP; bottom panels) cells for expression of KIT, TIE2, and FLK1 cell-surface markers. Each dot plot corresponds to the concatenation of the data from 3 individual E8.5 embryos of the indicated genotype using the concatenation function of FlowJo. Numbers indicate percentage. (B) Merged fluorescence and bright field images of late headfold E8.5 Gfi1b+/GFP embryos (5× objective). n indicates node; and ng, neural groove.
Gfi1 and Gfi1b are expressed at the onset of hematopoiesis in vivo. (A) FACS analysis of CD41+GFP− (wild type; top panels), CD41+Gfi1-GFP+ (Gfi1+/GFP; middle panels), and CD41+Gfi1b-GFP+ (Gfi1b+/GFP; bottom panels) cells for expression of KIT, TIE2, and FLK1 cell-surface markers. Each dot plot corresponds to the concatenation of the data from 3 individual E8.5 embryos of the indicated genotype using the concatenation function of FlowJo. Numbers indicate percentage. (B) Merged fluorescence and bright field images of late headfold E8.5 Gfi1b+/GFP embryos (5× objective). n indicates node; and ng, neural groove.
To assess the effect of combined Gfi1 and Gfi1b deficiencies on hematopoietic development, we crossed Gfi1+/GFP and Gfi1b+/GFP animals. Gfi1/Gfi1b double knockout (Gfi1GFP/GFP/Gfi1bGFP/GFP, thereafter referred to as KO/KO) embryos displayed an earlier embryonic lethality than Gfi1b single knockout (thereafter referred to as WT/KO) embryos, which die by E14.5.21 Indeed by E11.5 and E12.5, KO/KO embryos were delayed in their development relative to control littermates, contained fewer visible red cells, and appeared to be dying as indicated by their Hoechst staining (supplemental Figure 7, supplemental Table 1). At an earlier stage, E9.5 KO/KO embryos already displayed a reduction in visible red blood cells compared with control littermates (Figure 4), suggesting that the production of mature primitive erythrocytes might be affected in the double KO embryos. To investigate this possibility, we analyzed βH1 hemoglobin expression in E8.5 KO/KO embryos and examined their potential to generate primitive erythroid colonies in replating assays. We found a severely reduced level of βH1 globin expression in these embryos and a dramatic decrease in the numbers of erythroid colonies generated by them (supplemental Figure 8).
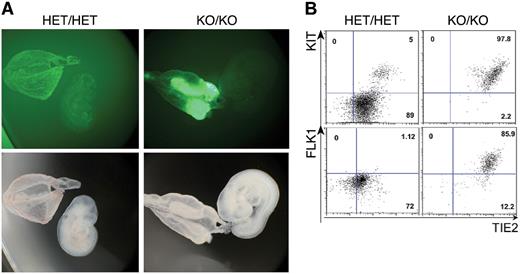
Loss of Gfi1 and Gfi1b blocks the release of blood cells from the yolk sac. (A) Fluorescence and bright field images of E9.5 HET/HET and KO/KO embryos and corresponding yolk sacs (2.5× objective). (B) FACS analysis of KIT/TIE2 expression among GFP+ cells of the indicated genotypes. Numbers indicate percentages.
Loss of Gfi1 and Gfi1b blocks the release of blood cells from the yolk sac. (A) Fluorescence and bright field images of E9.5 HET/HET and KO/KO embryos and corresponding yolk sacs (2.5× objective). (B) FACS analysis of KIT/TIE2 expression among GFP+ cells of the indicated genotypes. Numbers indicate percentages.
Under normal conditions, once the blood circulation is established by E8.25, hematopoietic cells produced in the yolk sac colonize the embryo proper.42,43 Accordingly, in E9.5 HET/HET embryos, the GFP+ cells—corresponding to CD41+ cells (Figure 4, supplemental Figure 9A)—were disseminated throughout the whole yolk sac and embryo proper (Figure 4). In contrast, in KO/KO embryos, the majority of GFP+ cells were only observed in restricted areas of the yolk sac and absent in embryo proper (Figure 4). Consistent with this result, GFP immunohistochemistry in E9.5 yolk-sac sections indicated that GFP+ cells were tightly packed together in KO/KO yolk sacs whereas in their HET/HET counterparts, GFP+ cells showed a more loose and individualized appearance (supplemental Figure 9B). One day later, in E10.5 embryos, GFP+ cells were more spread out in the KO/KO yolk sac, but this extraembryonic tissue was still only partially populated with GFP-positive cells and the embryo proper remained completely devoid of GFP-expressing cells (supplemental Figures 10-11, supplemental Video 3). This striking phenotype could be either explained by an inherent inability of the KO/KO cells to move, or alternatively by extrinsic factors such as a deficient vascular system or a lack of heartbeat. However, we observed that E10.5 KO/KO embryos have a normal heartbeat (supplemental Video 4), excluding a lack of circulation as the reason why the GFP+ cells do not move out of the yolk sac. Furthermore, when we performed immunohistochemistry staining for PECAM on these embryos we did not observe any vascular defects (Figure 5). Therefore, these findings suggest that the KO/KO cells are intrinsically unable to move. Finally, FACS analysis showed that at E9.5, most CD41+ cells in KO/KO embryos still expressed KIT and high levels of the endothelial markers TIE2 and FLK1, whereas in contrast, HET/HET CD41+ cells had mostly down-regulated the expression of these markers (Figure 4B, supplemental Figure 9A). Altogether these data indicate that in the combined absence of Gfi1 and Gfi1b, cells normally expressing these genes are unable to undergo changes in phenotype and morphology required for their dissemination into the yolk sac and the embryo.
KO/KO E10.5 embryos have a normal vascular system. Whole-mount CD31 staining of HET/HET (left) and KO/KO E10.5 embryos.
KO/KO E10.5 embryos have a normal vascular system. Whole-mount CD31 staining of HET/HET (left) and KO/KO E10.5 embryos.
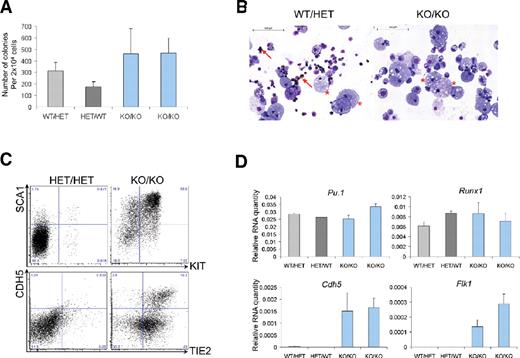
To evaluate whether these cells were also affected in their competence to generate hematopoietic cells, E9.5 KO/KO yolk sacs were dissociated into single cells and replated in clonogenic assays. Interestingly, once liberated from their original cell layer, these cells generated significant numbers of hematopoietic colonies (Figure 6A). Consistent with previous results (supplemental Figure 8 and Saleque et al21 ), we did not detect any erythroid cells in these colonies, but myeloid cells such as macrophages were clearly present (Figure 6B, supplemental Figure 12). Accordingly, hematopoietic genes such as Itgam (Cd11b), Runx1, Tal1, and Pu.1 were detected in cells of the KO/KO-derived colonies (Figure 6D, supplemental Figure 12B). The cells within these colonies expressed significant levels of the endothelial markers Sox7, Eng, Tie2, Cdh5, and Flk1 (supplemental Figure 12, Figure 6C-D). Additionally, the hematopoietic marker SCA144 was also highly expressed in KO/KO cells compared with normal hematopoietic colonies (Figure 6C). Yolk-sac cells from E10.5 KO/KO also generated hematopoietic colonies (supplemental Figure 13), whereas no colonies were generated from embryo-proper cells (supplemental Figure 13), a finding consistent with the complete absence of GFP+ cells in the embryo proper (supplemental Figures 10-11). Taken together, our data demonstrate that in Gfi1- and Gfi1b-deficient embryos, cells committed to a hematopoietic fate are generated but they maintain the expression of endothelial genes and are intrinsically unable to move freely and distribute into all embryonic tissues.
Cells lacking Gfi1 and Gfi1b are still able to generate hematopoietic cells. (A) Number of hematopoietic colonies generated by yolk-sac cells with the indicated genotypes after 8 days of culture. Values in the histogram correspond to an average of 3 technical replicates; the error bars correspond to the SEM. (B) Cytospins from hematopoietic colonies from panel A were stained with O-dianisidine, May-Grünwald-Giemsa. Arrows indicate erythroid cells (brown staining); stars indicate macrophages.(C) FACS analysis of SCA1/KIT and CDH5/TIE2 expression among cells of the indicated genotypes after clonogenic assay. Numbers indicate percentages. (D) Quantitative RT-PCR analysis of hematopoietic colonies generated in clonogenic assay.
Cells lacking Gfi1 and Gfi1b are still able to generate hematopoietic cells. (A) Number of hematopoietic colonies generated by yolk-sac cells with the indicated genotypes after 8 days of culture. Values in the histogram correspond to an average of 3 technical replicates; the error bars correspond to the SEM. (B) Cytospins from hematopoietic colonies from panel A were stained with O-dianisidine, May-Grünwald-Giemsa. Arrows indicate erythroid cells (brown staining); stars indicate macrophages.(C) FACS analysis of SCA1/KIT and CDH5/TIE2 expression among cells of the indicated genotypes after clonogenic assay. Numbers indicate percentages. (D) Quantitative RT-PCR analysis of hematopoietic colonies generated in clonogenic assay.
Discussion
In this study, we identified the GFI1 and GFI1B proteins as RUNX1 transcriptional targets at the endothelial to hematopoietic transition stage. We then demonstrated that GFI1 and GFI1B are able to trigger, in the absence of RUNX1, the down-regulation of endothelial markers and the formation of round cells, a morphologic change characteristic of the EHT. We also established that the repressive activity of GFI1 is critical to induce this change in morphology. GFI1 and GFI1B have been shown to suppress gene expression through binding to DNA and, subsequently, recruitment of corepressors such as histone deacetylases and (de)methylases leading to the removal of transcriptional activation marks and generation of repressive marks.45-47 Altogether, our findings suggest a scenario in which both GFI1 proteins suppress endothelial, and potentially other cell-adhesion molecules, gene expression during the transition from hemogenic endothelium to hematopoietic fate, leading to an epigenetic silencing of the endothelial signature in blood cells.
Although our study has clearly identified the 2 GFI proteins as main effectors driving the change from an endothelial to a hematopoietic morphology, they cannot on their own give full hematopoietic competence to the Runx1−/− hemogenic endothelial cells (Figure 7). These results suggest that hematopoietic specification from the hemogenic endothelium could be viewed as 2 RUNX1-dependent processes: (1) extinction of the endothelial cellular identity and (2) activation of the hematopoietic gene expression program. The 2 processes might be interrelated, as effectors implicated in the activation and function of the hematopoietic program could also participate in the loss of the endothelial identity and reciprocally. Indeed, our study identified the 2 GFI proteins as main effectors of endothelial identity loss but also indicated that they are able to induce, directly or indirectly, the expression of some early hematopoietic genes, although without supporting a full development of mature blood cells. The other downstream transcriptional target(s) of RUNX1 supporting the full commitment to a hematopoietic fate remains to be identified. The transcription factor PU.1 stands as a strong potential candidate for regulating hematopoietic genes because of its already well-established regulation by RUNX1 in early blood development.36 However, we observed that PU.1 on its own was not able to rescue the Runx1 KO deficiency and activate expression of hematopoietic genes (Figure 1C-D). These results suggest that other(s) effectors acting downstream of RUNX1 are required for the full acquisition of hematopoietic properties. Current efforts in the laboratory are aimed at identifying these factors.
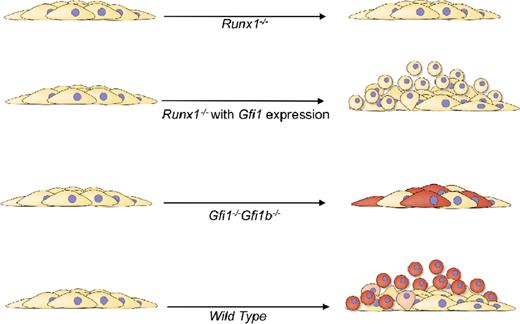
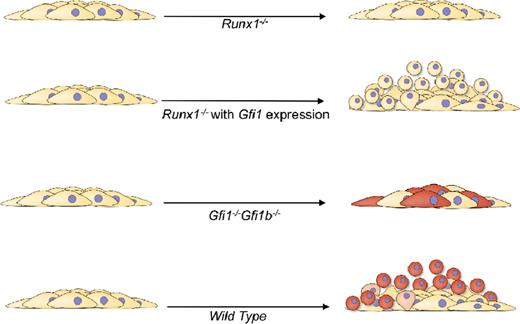
Model of the functional requirements for GFI1/GFI1b and RUNX1 during yolk-sac EHT. RUNX1 is required for both the change in morphology and acquisition of hematopoietic competence (cells turning red). GFI1/GFI1B are essential for the change in morphology, and additional unknown transcriptional targets of RUNX1 are required for acquisition of full hematopoietic competence.
Model of the functional requirements for GFI1/GFI1b and RUNX1 during yolk-sac EHT. RUNX1 is required for both the change in morphology and acquisition of hematopoietic competence (cells turning red). GFI1/GFI1B are essential for the change in morphology, and additional unknown transcriptional targets of RUNX1 are required for acquisition of full hematopoietic competence.
Our in vivo studies indicate that KO/KO embryos, dying on or before E11.5, suffer from an earlier and stronger phenotype than the single Gfi1 or Gfi1b KO embryos. These results are compatible with a model of functional compensation in blood development between both proteins in the single KO embryos. This concept is supported by the finding that both genes are expressed in the Tie2hiKit+FLK1+CD41+ cell population of E8.5 embryos, corresponding to cells becoming hematopoietic progenitors (Figure 3). The KO/KO embryos appear also to be dying earlier than Runx1−/− embryos dying by E12.5-E13.5. The Runx1−/− embryos are completely devoid of definitive hematopoietic precursors whereas earlier primitive erythroid development is relatively unaffected in these mice.27,40 One likely explanation for this earlier phenotype is therefore that GFI1 and GFI1B are critical for the development of functional primitive erythroid cells. The loss of Gfi1b has previously been shown to allow primitive erythrocyte production but these cells have an abnormal morphology and a delayed maturation.21 We found a severely reduced level of βH1 globin expression and a dramatic decrease in the numbers of erythroid colonies generated by E8.5 KO/KO embryos, suggesting that the additional absence of GFI1 led to more severe impairment of primitive erythropoiesis. The successful production of primitive erythroid cells in Runx1−/− embryos could be explained by the low levels of Gfi1 and Gfi1b expression in the absence of RUNX1 (Figure 1A).
The phenotype of the KO/KO embryos of a complete absence of hematopoietic precursors in the embryo proper has striking similarities with that of the Ncx1−/−48 and Rac1−/−49 mutant embryos. The Ncx1 gene encodes a sodium calcium exchanger and its absence leads to a lack of heartbeat and blood flow.50 It has been shown recently that in Ncx1−/− embryos, the hematopoietic progenitors produced in the yolk sac do not migrate to the embryo proper but remain normal functionally.48 Rac1 belongs to the Rho GTPase family and when specifically deleted in the hematopoietic lineage, yolk sac–derived blood progenitors are unable to migrate to the embryo proper.49 Collectively, these results suggest that up to E10.5, all blood progenitors detected in embryos originate from the yolk sac. However, the fact that Gfi1 and Gfib expression is detected at the very early onset of hematopoietic development from the hemogenic endothelium suggests that the lack of GFP+ cells observed in our study might also reflect an earlier defect at the level of hemogenic endothelium cells. This would suggest that either the development in situ of an intraembryonic hemogenic endothelium program requires GFI1(s) activity or that the intraembryonic hemogenic endothelium cell population originates from the yolk sac.
In conclusion, our results shed new light on the molecular and cellular mechanisms leading to the generation of the first blood cells. These findings have important implications not only for our understanding of hematopoietic development, but also for the design of novel experimental strategies that will permit the generation of blood cells for cell-replacement therapies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Catherine Stephenson, Stella Pearson, and the following facilities of the Paterson Institute for Cancer Research for technical support: Advanced Imaging, Biologic Resources Unit, Flow Cytometry, Histology and Molecular Biology Core Facility. They also thank Daniel Bilbao Cortes, head of the EMBL Monterotondo Flow Cytometry Facility, for assistance in this project.
This work was supported by grants from Cancer Research UK (G.L. and V.K.), the Biotechnology and Biologic Sciences Research Council (G.L., V.K., C.B.), the European Molecular Biology Laboratory (C.L.), and the European Community (FP6 Integrated Project EuTRACC, grant LSHG-CT-2007-037445).
Authorship
Contribution: C.L. and M.M. designed and performed the research, analyzed the data, and wrote the manuscript; M.S., R.P., M.L., G.C., Ö.V., and N.K.W. designed and performed experiments; T.M. contributed vital reagents; C.B. and B.G. designed and supervised research; and V.K. and G.L. designed and supervised the research project, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Georges Lacaud, Cancer Research UK Stem Cell Biology Group, Paterson Institute for Cancer Research, University of Manchester, Manchester M20 4BX, United Kingdom; e-mail: glacaud@picr.man.ac.uk.