Abstract
Among spliceosome component mutations, those involving SF3B1 are most frequent in myelodysplastic syndromes with ring sideroblasts (MDS-RS; ∼ 75% incidence) and SRSF2 in chronic myelomonocytic leukemia (∼ 28% incidence). We recently reported on the lack of prognostic significance for SF3B1 mutations in both MDS-RS and primary myelofibrosis (PMF). In the current study, we examined the prevalence and prognostic relevance of SRSF2 mutations in PMF. Among 187 patients screened, 32 (17%) harbored SRSF2 monoallelic mutations affecting residue P95. Significant associations were demonstrated between SRSF2 mutations and advanced age (P < .01), IDH mutations (P < .01), and higher DIPSS-plus risk category (P = .03). SRSF2 mutations were associated with shortened overall (P < .01) and leukemia-free (P < .01) survival; the adverse effect on survival was independent of DIPSS-plus (P = .01; HR = 1.9; 95% CI, 1.1-3.0) and IDH mutations (P < .01; HR = 2.3; 95% CI, 1.4-3.8). In conclusion, SRSF2 mutations are relatively common in PMF, cluster with IDH mutations, and are independently predictive of poor outcome.
Introduction
Genes encoding for different components of the RNA splicing machinery (SF3B1, SRSF2, U2AF1, U2AF65, ZRSR2, SF3A1, SF1, and PRPF40B) were recently shown to be recurrently mutated in a spectrum of myeloid malignancies1 ; cumulative mutational frequencies were ∼ 85% in myelodysplastic syndromes associated with ring sideroblasts (MDS-RS), 44% in MDS without RS, 55% in chronic myelomonocytic leukemia, 26% in therapy-related MDS or acute myeloid leukemia, 7% in primary acute myeloid leukemia, and 9% in myeloproliferative neoplasms. SF3B1 mutations were the most frequent (∼ 75%) in MDS-RS and SRSF2 mutations in chronic myelomonocytic leukemia (∼ 28%). SF3B1 mutations have also been described in chronic lymphocytic leukemia2 and, at a much lower frequency, in other solid tumors and cancer cell lines.3 SRSF2 mutations are also seen in other myeloid malignancies, including MDS, acute myeloid leukemia, myeloproliferative neoplasm and juvenile myelomonocytic leukemia.1,4 It is currently assumed that SF3B1, SRSF2, and other spliceosome-related mutations alter pre-mRNA splicing,5,6 but their exact mechanism of oncogenesis is not clear.
Current prognostication in primary myelofibrosis (PMF) is based on the Dynamic Prognostic Scoring System (DIPSS)–plus system that uses 8 clinical parameters, including karyotype7 ; furthermore, a specific group of cytogenetic abnormalities are associated with a particularly worse outcome.8 Such observations raise the possibility of using additional genetic markers to further refine prognostication in PMF. To that end, mutations involving IDH,9 EZH210 or ASXL111 have so far been associated with poor survival in PMF whereas those involving JAK2,12 MPL,13 TET2,14 or SF3B115 have not. In the current study, we screened 187 patients with PMF for SRSF2 mutations, to describe their prevalence, phenotypic correlates, and prognostic relevance in the context of DIPSS-plus and the presence or absence of IDH mutations.
Methods
The current study was conducted in accordance with the Declaration of Helsinki and approved by the Mayo Clinic Institutional Review Board. Patients were selected based on availability of archived DNA and bone marrow examination at time of sample collection for mutation screening. Diagnosis of PMF was according to the World Health Organization criteria, and patients with blast-phase disease were not included in the current study.16 Bone marrow pathology reports on SRSF2-mutated patients were rereviewed to look for the presence or absence of ring sideroblasts. Detailed analysis of clinical parameters and cytogenetic findings, at time of sample collection, was performed to risk stratify patients according to both DIPSS17 and DIPSS-plus.7 JAK2, MPL, IDH, and SF3B1 mutation screening was according to previously described methods.15,18-20 Standard statistical methods were used for parameter comparison, and survival curves were prepared by the Kaplan-Meier method and compared by the log-rank test. P < .05 was considered significant. The Stat View (SAS Institute) statistical package was used for all analyses.
SRSF2 mutation screening was performed using bone marrow aspirate- or granulocyte-derived DNA. To amplify SRSF2, a nested PCR approach was used in which 2 rounds of PCR were performed and focused around the P95 mutational hot spot (exon 1). Primers used are as follows: SRSF2 outer forward: 5′-CAAGGTGGACAACCTGACCT-3′; SRSF2 outer reverse: 5′-AGACGCCATTTCCCCAGT-3′; SRSF2 inner forward: 5′-GTGGACAACCTGACCTACCG-3′; SRSF2 inner reverse: 5′-CCTCAGCCCCGTTTACCT-3′. Cycling conditions were the same for both rounds and consisted of: an initial denaturation at 95°C for 10 minutes; 35 cycles of (denaturation at 95°C for 1 minute, annealing at 56°C for 1 minute, and extension at 72°C for 1 minute); and final extension at 72°C for 10 minutes. PCR products were purified after both rounds of PCR, and the final product was subjected to bidirectional sequence analysis.
Results and discussion
A total of 187 patients were studied, and their clinical and laboratory characteristics are listed in Table 1. Median age was 61 years, and 63% were males. DIPSS-plus risk distributions were 13% low, 13% intermediate-1, 37% intermediate-2, and 35% high-risk. Karyotype was available in all but one patient and was normal in 61% and showed either favorable (29%) or unfavorable (10%) abnormalities in the remaining. All 187 study patients were fully annotated for JAK2, 182 for MPL, 184 for IDH, and 165 for SF3B1 mutations; mutational frequencies were 59%, 6%, 8%, and 7%, respectively. Treatment included splenectomy in 28 (15%) patients and allogenic stem cell transplantation in 11 (6%; Table 1). Other therapies were also at the discretion of the treating physician and included noncytoreductive agents, such as androgen preparations, prednisone, erythropoiesis-stimulating agents, thalidomide, anagrelide, and cytoreductive agents, such as hydroxyurea, interferon-alfa, and lenalidomide. Less than 5% of the patients received JAK inhibitor therapy. To date, 117 (63%) deaths and 31 (17%) leukemic transformations have been recorded.
Thirty-two patients (17%) harbored SRSF2 mutations, the majority of which were missense (12 P95H, 8 P95L, 4 P95R, and 1 P95S) but included 6 patients with a 24-bp deletion (delP95)–R102 (PPDSHHSR) and 1 patient with 274-275insACC (G93D;P95R). As illustrated in Table 1, SRSF2 mutations clustered with both IDH1 (P < .01) and IDH2 (P < .01) mutations and a borderline association was also seen with JAK2V617F (P = .11). In contrast, none of the SRSF2-mutated patients expressed MPL mutations, whereas one patient expressed both SRSF2 and SF3B1 mutations. In a subset of 23 SRSF2-mutated patients in whom bone marrow iron stains were performed, ring sideroblasts were seen in 3 patients; SF3B1 mutation screening was negative in all these 3 patients. Iron stains were not available in the sole SFSR2—SF3B1 double-mutant patient. Among SRSF2-mutated patients, comparison of IDH-mutated versus IDH-unmutated and JAK2-mutated versus JAK2-unmutated cases suggested significant associations for SRSF2-JAK2V617F double mutants with younger age, higher leukocyte count, higher hemoglobin profile, and lower prevalence of IDH mutations (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Consistent with the latter observation, SRSF2-IDH double mutants were less likely to express JAK2V617F (supplemental Table 1).
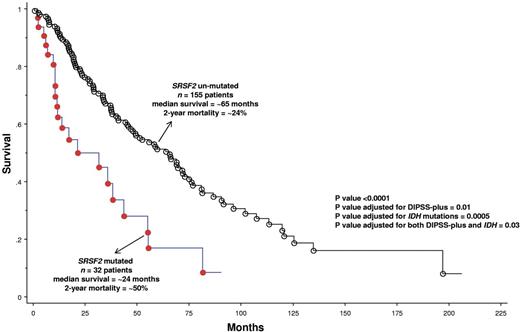
The presence of SRSF2 mutations was significantly associated with advanced age (P < .01) and high-risk category per DIPSS-plus (P = .03; Table 1). Most importantly, SRSF2 mutations were associated with shortened overall (P = .0001; HR = 2.6; 95% CI, 1.6-4.2; Figure 1) and leukemia-free (P = .0004; HR = 4.3; 95% CI, 1.9-9.6; supplemental Figure 1) survival; the adverse effect on survival was independent of DIPSS (P = .003; HR = 2.1; 95% CI, 1.3-3.5), DIPSS-plus (P = .01; HR = 1.9; 95% CI, 1.1-3.0) or the presence of IDH mutations (P = .0005; HR = 2.4; 95% CI, 1.5-3.9). The adverse effect on leukemia-free survival was independent of karyotype (P = .0002; HR = 4.9; 95% CI, 2.2-11.3) and IDH mutations (P = .003; HR = 3.6; 95% CI, 1.6-8.4). We have previously shown an adverse prognostic effect from mutant IDH, in PMF,9 and the current study suggests that mutant SRSF2 is independent of both IDH mutations and DIPSS-plus in its prognostic relevance.
Overall survival of 187 patients with PMF (median survival = 55 months; 2-year mortality = 28%) stratified by the presence or absence of SRSF2 mutations.
Overall survival of 187 patients with PMF (median survival = 55 months; 2-year mortality = 28%) stratified by the presence or absence of SRSF2 mutations.
Our observations are particularly akin to those of Thol et al who recently analyzed SRSF2, U2AF1, ZRSR2, and SF3B1 mutations in a cohort of 193 MDS patients.21 As was the case in our PMF patients, SRSF2 mutations in their MDS patients (mutational frequency of 12%) clustered with IDH1mutations and predicted inferior overall and leukemic-free survival, whereas SF3B1 mutations had no impact on survival. Similarly, Zhang et al have recently reported an adverse outcome in SRSF2-mutated blast-phase myeloproliferative neoplasm (∼ 19% mutational frequency),22 and Damm et al reported an association between mutant SRSF2 and MDS with excess blasts.23 Taken together, these observations identify SRSF2 mutations as possibly the most important spliceosome-related mutation; in terms of utility as a prognostic marker, its value in this regard is further enhanced by its relatively common occurrence. Finally, other investigators have identified EZH2 and ASXL1 mutations as having a negative prognostic impact in PMF, but their value in the context of DIPSS-plus remains to be clarified.10,11 Regardless, taken together, these observations contribute to our insight regarding leukemic transformation in PMF and also suggest the possibility of incorporating molecular markers into currently established prognostic scoring systems.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.T. designed and performed the research, contributed patients, performed the statistical analysis and its interpretation, and wrote the manuscript; T.L.L. designed and performed the research, analyzed the data, and contributed to writing the manuscript; T.J. abstracted and analyzed data; C.M.F. performed the experiments; M.P. contributed patients and helped design the research; C.A.H. reviewed pathology; R.P.K. reviewed cytogenetic findings; A.P. designed research, contributed patients, and helped interpret data; and all authors approved the final draft of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ayalew Tefferi, Mayo Clinic, 200 First St SW, Rochester, MN 55905; e-mail: tefferi.ayalew@mayo.edu.