Abstract
Abstract 1398
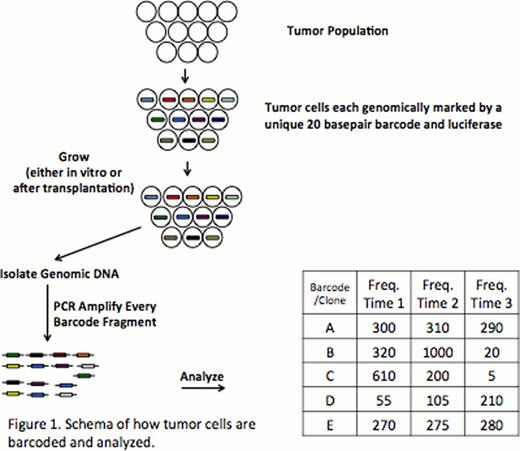
Acute myeloid leukemia (AML) is a heterogeneous malignancy that continues to have a significant relapse rate in both adult and pediatric patients. This heterogeneity manifests itself between patients because different patients will have different gene mutations causing their AML (inter-patient heterogeneity). It also manifests as intra-patient heterogeneity as within a single patient there are different sub-clones that contribute differentially to primary disease and relapse. For example, relapse after chemotherapy can occur from a genetically distinct sub-clone that presumably was relatively resistant to the administered chemotherapy. The dynamics of clonal relapse and the contribution of intra-patient functional heterogeneity (of either genetic or epigenetic origin), however, remains poorly understood. We have developed an assay using a molecular barcode system and next generation sequencing to simultaneously track the fate of ∼14,000 different clones either in vitro or in vivo (Figure 1). Individual clones can therefore be tracked over time even in complex populations. We validated this assay on a human AML cell line, THP1, over multiple population doublings, describe how the heterogeneity can be monitored and quantified over time and describe that even in this cell line the population continues to undergo dynamic clonal changes.
A lentiviral barcode library consisting of ∼14,000 unique 20 basepair sequences (the molecular barcode) was transduced into THP1 cells at a multiplicity of infection of 0.05 (to assure that each cell only acquired a single barcode). Barcoded cells were purified by selection for GFP by flow cytometry. Cells were maintained in RPMI at a concentration of 2×106 cells/ml in triplicates and passaged every 2∼3 days. The percentage of GFP expression was assessed every 30 population doublings (PD). Cells were passaged to a maximum of 90 population doublings. Genomic DNA was extracted every 10 population doublings (PD), barcodes were amplified by PCR using primers bracketing the barcode region to produce a 250 bp product, and gel purified 250 bp products were submitted to Illumina™ high throughput sequencing to quantify the abundance of each barcode in the population. Barcodes were clustered allowing for a minimum quality score of 30 and a maximum of 1 mismatch. Clusters were ordered and plotted by frequency of occurrence from highest to lowest. The frequency of barcode clusters at specific PD was compared between the triplicates.
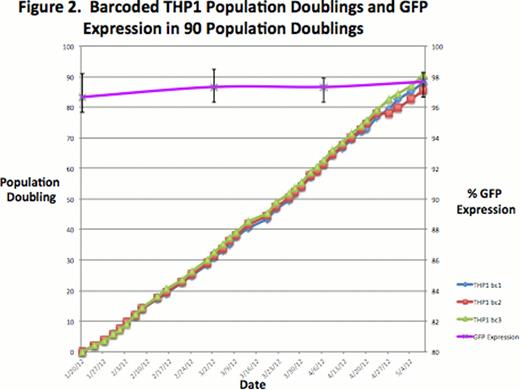
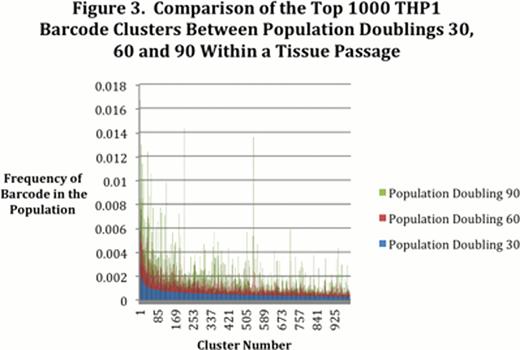
The population doublings and percentage of GFP expression of barcoded THP1 cells was nearly identical in the triplicate cell passage of up to 90 population doublings (Figure 2. THP1 bc1 = barcoded THP1 tissue passage 1 of triplicate, THP1 bc2 = barcoded THP1 tissue passage 2 of triplicate, THP1 bc3 = barcoded THP1 tissue passage 3 of triplicate). A high fraction of these barcoded cells (> 90%) expressed GFP consistently throughout the 90 population doublings. On the other hand the heterogeneity, represented by clusters of barcodes in a single cell passage at PD 30, 60 and 90, varied between PD as shown in the stacked histogram (Figure 3). PD 30 was the reference PD and was sorted from highest cluster frequency to lowest; all other PDs were compared to PD 30.
Subpopulations of cells are dynamic even in established cell lines, including the THP1 human AML cell line. Fluctuations in barcode frequencies were observed between PD within a sequential cell passage. A high degree of heterogeneity was therefore maintained and thus represents the highly dynamic nature of this AML cell line. This barcode assay can be a powerful tool to assess how population heterogeneity can be affected by environmental pressures such as chemotherapy. Our future effort is directed at challenging barcoded THP1 cells with cytarabine, a foundation of AML therapy, to determine how chemotherapy affects the functional heterogeneity of a dynamic heterogeneous population. In addition, we are also assessing how heterogeneity is influenced in vivo by first transplanting barcoded THP1 cells into NSG mice and then challenging the transplanted mice with chemotherapy.
Hsu:Child Health Research Institute and the Stanford CTSA (grant no. UL1 RR025744): Research Funding; Rosa Wann and Marjorie Shannon Fellowship: Research Funding. Porteus:Alex's Lemonade Stand Foundation: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.