Abstract
Abstract 2906
Chronic Lymphocytic leukemia (CLL) is currently incurable using conventional therapies. CLL cells can evade killing by various therapeutic strategies. However the precise mechanisms are currently unknown. Autophagy is regulated by a complex system of proteins, and is used by both normal and malignant cells as a protective mechanism against cellular stress induced by starvation, hypoxia, reactive oxygen species (ROS) and endoplasmic reticulum (ER) stress. In malignant cells autophagy was shown to promote tumorigenesis and/or resistance to chemotherapy. Therefore we hypothesized that autophagy may play a role in CLL biology.
Autophagy can also promote cell death when stress signals are elevated above a particular threshold for a prolonged period of time. In this study we investigated the basal expression levels of autophagy specific genes and the effect of autophagy specific inhibitors (Bafilomycin, 3-methyladenine and hydroxychloroquine) and inducers (Phenethyl isothiocyanate) on CLL survival. Phenethyl isothiocyanate (PEITC) is about to enter clinical trials for CLL (NCT00968461).
We have investigated induction of components of the autophagic pathway following treatment of CLL cells in vitro with a range of chemical inhibitors. Immunoblotting was carried out to investigate components of the autophagy pathway using phosphorylation state-specific and pan-reactive antibodies. Bafilomycin (BAF), 3-methyladenine (3-MA) and hydroxychloroquine (HCQ) toxicity towards CLL samples were evaluated by Annexin V/PI staining, MTT assay and immunoblotting for cleavage of the caspase 3 substrate poly(ADP ribose) polymerase (PARP) from its 116KDa to its 85KDa form. PEITC was used at concentrations between 2.5 and 25μM to investigate its effect on signaling. Autophagy was quantitated by immunoblotting of LC3-I and LC3-II. Lipidation of LC3 from LC3-I to LC3-II is a surrogate marker of autophagy and is essential for autophagasome formation. Immunoblotting was also performed for ATG3, ATG5 and ATG7, key components of the autophagy pathway. Monodansylcadaverine (MDC) was used with immunofluorescence and FACS analysis to investigate increases in autophagasome formation. Transmission electron microscopy (TEM) was used to confirm double membrane bound autophagosomes. Co-immunoprecipitation was used to evaluate if Beclin-1 was sequestered by Bcl-2 preventing autophagy. Its release from Bcl-2 enables Beclin-1 to interact with other autophagy specific proteins and initiates autophagasome formation.
LC3-I was lipidated to LC3-II (p=0.019) and ATG3 (p=0.021) was upregulated to a greater extent in CLL samples compared with normal B-cell controls at basal levels. This suggested that autophagy was active to a greater extent in CLL samples compared with normal individuals. In addition Beclin was dissociated from Bcl-2 in CLL samples indicating that autophagy was active. Autophagy appears to be a pro-survival mechanism in untreated CLL cells as inhibiting basal levels of autophagy with autophagy inhibitors BAF (50–200nM), 3-MA (5–10mM) and hydroxychlorquine (5–10μM) resulted in CLL apoptosis as shown by MTT, Annexin V/PI analysis and PARP cleavage.
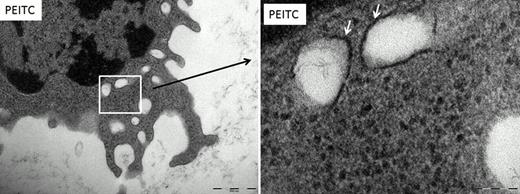
Interestingly augmenting autophagy was also capable of inducing apoptosis in CLL samples. Treatment with PEITC caused an increase in punctate staining using MDC which is suggestive of autophagosome formation. We went on to determine that PEITC further induced LC3-II lipidation using immunoblotting and showed a substantial increase in overall LC3 protein expression. PEITC also induced the expression of ATG3, a key protein in the autophagy pathway. We then evaluated autophagosome formation using TEM (Figure 1). Our data showed greater numbers of autophagosomes in the PEITC treated samples compared to the untreated controls.
Therefore autophagy in CLL sits on a knife-edge, such that perturbations that either increase pro- death or decrease pro-survival autophagy signals can result in CLL cell death, depending on the duration and intensity of the signal.
Transmission electron microscopy of CLL cells
CLL cells were treated with 10μM PEITC. Double membrane bound organelles were found in the CLL cells after treatment which were not present in the no addition control (depicted by the arrows). These organelles are autophagsomes. Magnification (left picture) ruler is 500nM, (right picture) ruler is 100nM
Transmission electron microscopy of CLL cells
CLL cells were treated with 10μM PEITC. Double membrane bound organelles were found in the CLL cells after treatment which were not present in the no addition control (depicted by the arrows). These organelles are autophagsomes. Magnification (left picture) ruler is 500nM, (right picture) ruler is 100nM
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.