Abstract
Abstract 319
Outcome in multiple myeloma (MM) is heterogeneous and the assessment of individual risk based on molecular features is essential to make personalised therapy possible. Epigenetic modifications contribute to individual tumour biology and, consequently, clinical behaviour. Here, we present data on the association between differential methylation, gene expression and survival in 161 newly-diagnosed MM patient samples. The aim of the study is to provide novel insights based on genome wide data into the epigenetic biology of myeloma as well as to define prognostically relevant epigenetic markers.
Methylation data from Illumina Infinium HumanMethylation27 BeadChips for 161 presentation MM samples was used for the analysis. All patients had been treated in the MRC Myeloma IX trial with a median follow-up of 5.9 years. FISH data was available for 144 patients and Affymetrix hgU133 plus 2.0 gene expression data was available for 117 patients. Methylation values were binarised using cut-off values of 30% (hypomethylated) or 70% (hypermethylated) methylation. Multivariate Cox regression analyses were conducted on the 27,578 methylation probes to investigate their association with overall survival (OS). Multiple testing correction of results using the Benjamini-Hochberg algorithm with a 5% FDR was performed.
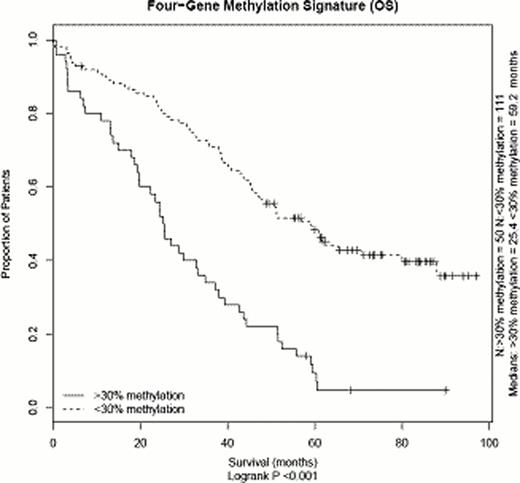
Using a logistic regression model testing, we defined genes for which differential methylation was associated with differences in overall survival (OS). In the group defined by a 30% cut-off value, 260 probes (equivalent to 247 genes), were statistically associated with OS differences as well as 127 probes (124 genes) in the 70% cut-off value group. Gene set enrichment analysis using KEGG and PathwayCommons datasets revealed statistical overrepresentation of metabolic pathways (n=15), regulation of IGF (n=3) and integrin/focal adhesion (n=6). We then looked for genes for which methylation and expression were correlated. For 19 genes, gene expression differed significantly between the samples with <30% vs ≥30% methylation (Wilcoxon p<0.05). Of these, four candidate genes with known tumour gene properties were selected for further analyses. Expression for these genes showed significant inverse correlation with methylation. OS survival was significantly lower in the methylated (≥30%) vs the unmethylated (<30%) fraction for each of the genes, with 22.1 (n=20) vs 50.9 months (n=141), 24.9 (n=12) vs 47.7 months (n=149), 19.7 (n=13) vs 46.6 months (n=148) and 24.4 (n=30) vs 48.3 months (n=131) [log-rank p<0.05 for all genes]. Hypermethylation of two of the genes was associated with presence of the t(4;14) translocation, whereas it was not associated for the other two (Fisher's exact t-test). However, in multivariate analyses including the known risk factors t(4;14), del(17p), gain(1q), ISS and age, three genes retained statistical significance (Hazard ratios [HR] of 3.0 [CI 1.3–7.0], 6.1 [2.3–16.5] and 2.6 [1.5–4.8], resp.). When tested only in the subgroup of patients with presence of at least one adverse cytogenetic lesion, methylation status of three of the four genes discriminated patients with significantly shorter OS. A methylation signature was defined by presence of ≥30% methylation for one or more of the four genes, encompassing 50 patients. Survival of the patient group defined by the signature was significantly shorter (OS 25.4 vs 59.2 months, p<0.05) and the difference in survival retained statistical significance in a multivariate analysis (HR 3.4 [CI 1.8–5.4]).
In the present study we have analysed the association between gene methylation, expression and survival on a genome-wide scale. The candidate genes identified play important, non-overlapping roles in different aspects of the hallmarks of cancer, including cell cycle control, transcription/proliferation control, apoptosis and migration. These genes have not been identified in other GEP defined prognostic signatures and are independent of GEP signatures such as those defined by ourselves, UAMS or the Dutch group. The association between methylation and expression makes these genes promising targets for epigenetic therapies. Diagnostic tests for such methylation changes may allow the identification of patients in low and high risk disease subgroups.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.