In this issue of Blood, Tiacci et al uncover molecular survival mechanisms of the malignant Hodgkin-Reed-Sternberg cell (HRS) in a disease whose pathophysiology has until recently remained a mystery.1
Classical Hodgkin lymphoma (CHL) treatment was once the paragon of innovation in cancer. Therapeutic revolution came early for this disease: clinical outcomes improved drastically with the introduction of combination chemotherapy in the 1970s when 5-year survival rates for advanced disease improved from less than 30% to more than 60%. Improved supportive care, intensity-modified chemotherapy and radiotherapy schedules,2 and myeloablative salvage strategies have improved this to 80% to 90% today.
However, unlike the treatment revolution in chronic myeloid leukemia, where understanding of molecular pathophysiology (aberrant fusion protein BCR-ABL) facilitated targeted therapy (with tyrosine kinase active site-specific inhibitors), there has been no breakthrough in a molecular understanding of CHL yielding targetable pathways, or risk-stratifying prognostic markers. With the exception of HRS-expressed CD30-targeting toxin-conjugated antibody therapy brentuximab vedotin, application of novel therapies has made little impact on conventional therapy-refractory patients. Even the origin of the malignant cell has only recently been confirmed, with infectious and nonlymphoid etiology being postulated until the demonstration of B-cell receptor rearrangement and somatic hypermutation in the HRS confirmed its B-lymphocyte origin.3 The problem lay in studying a rare malignant cell overwhelmed by its abundant microenvironment.
Earlier work has shown that apoptosis pathway dysregulation seen in other hematologic malignancies, such as FAS, caspases, and BCL2, are rarely encountered in CHL, although MDM2 gains and TP53 mutations are relatively frequent (reviewed in Küppers4 ). Components of the paradigm survival pathways of normal and malignant lymphocyte physiology, nuclear factor κ-light chain enhancer of activated B cells (NF-κB) and Janus-associated kinase/signal transducer and activator of transcription (JAK/STAT), are frequently dysregulated in HRS cells but none are pathognomonic. Clearly many survival pathways are at work, some arising from the malignant cell, others delivered by the microenvironment.4 Notably, and unlike the non-Hodgkin lymphomas (NHLs), recurrent chromosomal rearrangements or specific genetic lesions have not been identified for CHL until recently, and remain the exception rather than the rule.
Previous gene expression profiling (GEP) studies have analyzed cell lines, which may be unrepresentative of primary HRS cells, or whole tumor, in which HRS cell mRNA contribution is diluted out by microenvironment. Findings have been inconsistent between research groups.5-7 Array comparative genomic hybridization studies of micro-dissected HRS have demonstrated copy number aberrations alluding to the genes responsible for HRS survival.8 A recent study of 29 micro-dissected cases found evidence of molecular subtypes of CHL relating to B-cell phenotype retention and cytotoxic T-cell molecule expression, while demonstrating macrophage-associated molecule CSF1R in the poorer prognosis subgroup.9
Tiacci et al have applied cutting-edge technology with laser-capture micro-dissection HRS-enrichment and comprehensive chip-based gene expression analysis to a substantial number of primary CHL cases. Essentially, and uniquely, the GEP data generated have been compared with carefully selected controls, nonmalignant germinal center B cells at various stages of maturation, other mature B cell–derived malignant cells, and, importantly, the only ex vivo model system for HRS investigation available at present: CHL-derived continuous cell lines. Bioinformatic analysis has thus yielded HRS-defining patterns of gene expression.
One key finding is the remarkable difference in GEP of the primary HRS cell, isolated fresh from its supportive microenvironment, compared with the autonomously growing cell lines: of profound importance to those who seek to generalize functional data derived only from cell lines. Overrepresentation of NF-κB target genes and suppression of B-cell programming is confirmed in both primary and cultured cells, but 2000 genes are differentially expressed, and unsupervised hierarchical clustering delineates primary CHL from cell lines to a greater extent than from other malignancies.
Despite emerging evidence that nodular lymphocyte-predominant Hodgkin lymphoma (NLPHL) and CHL are biologically quite distinct, while primary mediastinal B-cell lymphoma (PMBL) and CHL are related,10 this study finds more GEP similarities at the level of malignant cell between CHL and NLPHL than PMBL. Stratifying by histologic subtype, nodular sclerosis CHL-derived HRS cells map more closely to PMBL, while mixed cellularity CHL-derived cells resemble NLPHL. Surprisingly few GEP differences emerge based on EBV status, questioning the central role of the virus in producing the characteristic molecular changes in CHL, or alternatively demonstrating that either endogenous genetic mutations or EBV-transformation can induce a similar CHL-characteristic GEP. The cell of origin of CHL remains elusive after this study with very little overlap with GEP of plasma cells or germinal center B cells.
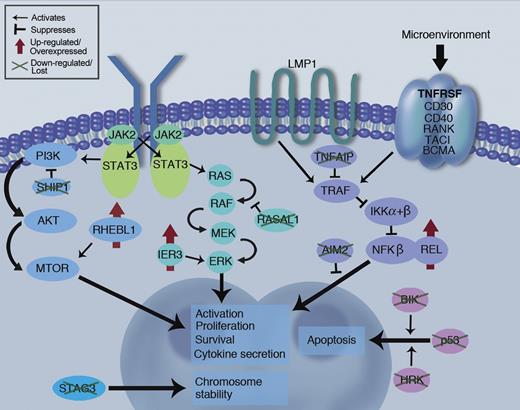
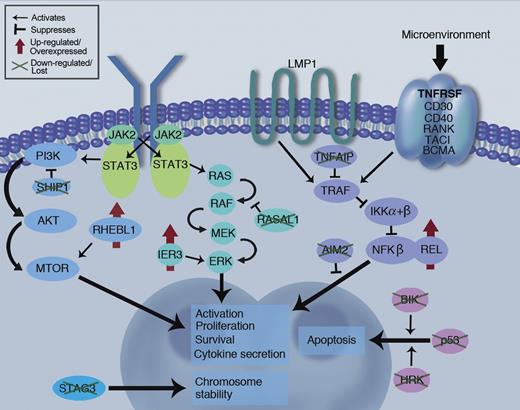
The holy grail of GEP work is the discovery of unique molecular drivers for a malignancy that will lead to novel, HRS-specific potentially drugable targets. This study has, for the first time, demonstrated several such up-regulated targets including RHEBL1 and IER3, molecules acting upstream of key survival mediators NF-κB and ERK. Notably, BIK, a proapoptotic factor, and SHIP1, a PI3-kinase inhibitor, are down-regulated in the HRS (see figure).
Schematic of key survival pathways at work in the HRS malignant cell. Novel findings of pathway dysregulation reported in Tiacci et al are highlighted (green crosses and red arrows), along with the established REL and P53 abnormalities already known from the literature. LMP1 signaling is likely a key survival pathway in EBV positive disease, while signaling through TNFRSF pathway may provide redundancy for this pathway in EBV negative disease, along with the JAK/STAT and PI3K/MTOR pathways. Professional illustration by Marie Dauenheimer.
Schematic of key survival pathways at work in the HRS malignant cell. Novel findings of pathway dysregulation reported in Tiacci et al are highlighted (green crosses and red arrows), along with the established REL and P53 abnormalities already known from the literature. LMP1 signaling is likely a key survival pathway in EBV positive disease, while signaling through TNFRSF pathway may provide redundancy for this pathway in EBV negative disease, along with the JAK/STAT and PI3K/MTOR pathways. Professional illustration by Marie Dauenheimer.
Guidance of therapeutic strategy or prognostication by stratifying patients using molecular markers has not been achieved for CHL, despite the promise shown in DLBCL with the emerging ABC/GCB dichotomy. This study provides a tantalizing suggestion of at least 2 distinct molecular subtypes, defined by GEP signatures of 3 proto-oncogenes, that had not been suspected as contributing to CHL. Unsupervised clustering discriminated 2 groups discriminated not by histologic subtype or EBV status, but rather by GEP signatures suggesting MYC, IRF4 and NOTCH1 activation, factors with well described roles in Burkitt lymphoma, myeloma, and T-cell acute lymphoblastic leukemia, respectively.
This important work of Tiacci et al has therefore advanced our understanding of the molecular drivers of CHL, uncovered pathways that may yield therapeutic targets and prognostic indicators and provided a gene signature explaining some of the biologic diversity of disease as well as potential unique markers for the proposed, but as yet unconfirmed CHL stem cell.11 With the caveat that the relatively small patient numbers and limited protein-level validation should be interpreted with some caution, it seems that a solution to the enigma of CHL comes closer still.
Conflict-of-interest disclosure: The authors declare no competing financial interests. ■