Abstract
C1q modulates the differentiation and function of cells committed to the monocyte-derived dendritic cell (DC) lineage. Because the 2 C1q receptors found on the DC surface—gC1qR and cC1qR—lack a direct conduit into intracellular elements, we postulated that the receptors must form complexes with transmembrane partners. In the present study, we show that DC-SIGN, a C-type lectin expressed on DCs, binds directly to C1q, as assessed by ELISA, flow cytometry, and immunoprecipitation experiments. Surface plasmon resonance analysis revealed that the interaction was specific, and both intact C1q and the globular portion of C1q bound to DC-SIGN. Whereas IgG reduced this binding significantly, the Arg residues (162-163) of the C1q-A chain, which are thought to contribute to the C1q-IgG interaction, were not required for C1q binding to DC-SIGN. Binding was reduced significantly in the absence of Ca2+ and by preincubation of DC-SIGN with mannan, suggesting that C1q binds to DC-SIGN at its principal Ca2+-binding pocket, which has increased affinity for mannose residues. Antigen-capture ELISA and immunofluorescence microscopy revealed that C1q and gC1qR associate with DC-SIGN on blood DC precursors and immature DCs. The results of the present study suggest that C1q/gC1qR may regulate DC differentiation and function through the DC-SIGN–mediated induction of cell-signaling pathways.
Introduction
C1q (460 kDa) is a collagen-like, hexameric glycoprotein predominantly synthesized by macrophages and dendritic cells (DCs).1,2 It belongs to the TNF and collectin (collagen-containing lectin) families of molecules, which contain collagen-like sequences contiguous with noncollagen-like stretches.3 Unlike most collectins, C1q does not contain a consensus carbohydrate recognition domain; instead, it contains sequences in its globular heads that allow it to bind to protein motifs in IgG or IgM.4 Therefore, C1q can bind to immune complexes and engage in complement-mediated microbial killing and phagocytosis.5 In addition, C1q is highly positively charged,6 which allows it to interact through ionic bonds with a multitude of negatively charged molecules.7
Whereas the role of C1q in the initiation of the classic complement pathway is well known, evidence has emerged indicating its involvement in numerous immunoregulatory processes, including removal of self-waste, regulation of T-cell proliferation, monocyte migration, and DC activation.8-11 C1q is composed of 2 major structural and functional domains: 6 globular “heads” (gC1q) linked to a collagen-like “stalk” (cC1q) or “tail.”12,13 Each of these domains binds to a ubiquitously expressed receptor: gC1qR binds to gC1q14 and cC1qR (calreticulin) binds to cC1q.15 Although both gC1qR and cC1qR lack a consensus motif for a transmembrane segment, each has been shown to recruit signaling partners with transmembrane domains. For example, some putative signaling partners for cC1qR include CD91 on monocytes,16 scavenger receptor A on APCs,17 CD59 on neutrophils,18 α2β1 integrin and glycoprotein VI on resting platelets,19 MHC class I on T cells,20 and CD69 on human PBMCs.21 Potential signaling partners for gC1qR are β1-integrins and CD44 on endothelial cells,22 vasopressin V2 receptor on the HEK 293 cell line, and the α(1B)–adrenergic receptor on the COS 7 cell line.23 Indications that C1q may function as an autocrine signal in DCs and regulate both innate and adaptive properties have emerged recently.8,9,24,25 However, whereas both C1q receptors are present on the surface of immature DCs (iDCs), little is known about their possible signaling partners on these cells.
iDCs, but not mature DCs, are a primary source of active C1q.1,26 Human monocyte-derived iDCs express high levels of both gC1qR and cC1qR, and their surface levels decrease in a maturation-dependent manner. Recently, SIGN-R1 has been shown to bind directly to C1q on DCs and to assemble C3 convertase without the traditional requirement for either Ab or factor B.27 SIGN-R1, which is expressed abundantly on the medullary and subcapsular macrophages in murine lymph nodes, is a homolog of the DC-specific ICAM-3–grabbing nonintegrin (DC-SIGN/CD209), which is expressed by human DCs.28,29 Therefore, we hypothesized that DC-SIGN may interact with C1q on human DCs and function as a transmembrane partner for C1q receptors.
In the present study, we present evidence that DC-SIGN not only binds to gC1q directly and specifically, but also that C1q and gC1qR associate with DC-SIGN on iDCs and blood DC precursors, indicating that C1q/gC1qR may regulate DC differentiation and function through DC-SIGN.
Methods
Reagents
The following reagents were used: Lymphoprep (Axis-Shield); BIAcore CM5 sensor chip (GE Healthcare); cell lysis buffer (Cell Signaling Technology); C1q-, C3-, and factor H–depleted human sera (CompTech); gC1qR and synthetic C1q-A peptides (GenScript); penicillin/streptomycin and RPMI 1640 medium (Gibco-Invitrogen); heat-inactivated FBS (HyClone Laboratories); DC-SIGN (Novus Biologicals); human recombinant GM-CSF and human recombinant IL-4 (PeproTech); p-nitrophenyl phosphate, EZ-Link Sulfo-NHS-LC-Biotin, and SuperSignal West Pico ECL reagent (Pierce); BSA, mannan, human IgG, and DAPI (Sigma-Aldrich); and Immu-Mount and neutravidin Ultralink resin beads (Thermo Fisher).
Abs used were: mAb and polyclonal Ab (pAb) against C1q (Quidel); mAb against DC-SIGN (eBiosciences); PE-conjugated goat anti–rabbit IgG F(ab′)2, FITC-conjugated sheep anti–rat IgG F(ab′)2 (Invitrogen); HRP-conjugated rabbit anti–goat IgG and donkey anti–rabbit IgG (GE Healthcare Bio-Sciences); and alkaline phosphatase (AP)–conjugated rabbit anti–goat IgG, donkey anti–rabbit IgG, and streptavidin-AP (Pierce).
Because of the sensitivity of DCs to endotoxin, highly purified and endotoxin-poor reagents and proteins were used.
Other proteins and Abs against gC1qR
Generation of monocyte-derived DCs
Monocyte-derived DCs were generated according to previously published methods.9 Briefly, PBMCs were purified from whole blood using Lymphoprep density gradient centrifugation following the manufacturer's instructions. Monocytes were further purified by adhesion selection on polystyrene plates (1 hour at 37°C) to ≥ 95 purity, as assessed by scatter profiles and microscopic observations. The cells were seeded at a final concentration of 0.5 × 106 cells/mL in RPMI medium containing 10% FBS, 100 U/mL of penicillin/streptomycin, 50 U/mL of human recombinant GM-CSF, and 50 ng/mL of human recombinant IL-4 to generate monocyte-derived DCs and grown for 4 days. The cells were harvested daily for analysis by flow cytometry. All reagents and cell-culture conditions were kept low in endotoxin. Viability of cells was typically ≥ 95% as assessed by Trypan blue exclusion. The research was approved by the Stony Brook University institutional review board and all human participants donating blood gave written informed consent in accordance with the Declaration of Helsinki.
Flow cytometry
Cells were collected and washed in PBA buffer (PBS containing 1% BSA and 0.01% NaN3). Nonspecific binding was blocked with 1 mg/mL of human IgG in PBA plus 1 × 106 cells (30 minutes at 4°C) and primary Abs or the appropriate isotype-matched controls were added to the cells (30 minutes at 4°C). Cells were washed twice in PBA, and further incubated with fluorescent-labeled secondary Abs (30 minutes at 4°C). The cells were washed in PBA, fixed in 1% formalin, and analyzed by with a FACSCalibur flow cytometer (BD Biosciences). For each analysis, 5000 events were collected. The data were analyzed using CellQuest Pro Version 5.1 software (BD Biosciences).
ELISA
Microtiter plates (MaxiSorb; Nunc) were coated with protein or Ab (5 μg/mL) in coating buffer (100mM Na2CO3/NaHCO3, pH 9.6; 2 hours at 37°C). Alternatively, synthetic C1q peptides were coated at 1.4 × 10−8M. After incubation, the wells were washed with PBS containing 0.5% NaCl and blocked with 3% BSA/PBS (1 hour at 37°C). This was followed by incubation with 5 μg/mL of protein or 100 μg of day 3 DC lysates in PBS containing 0.05% Tween or in PBS containing 10mM EDTA (1 hour at 37°C) for studying Ca2+-dependent binding. The wells were washed and incubated with primary Ab or appropriate isotype-matched controls (1 hour at 37°C). The bound proteins were probed with AP-conjugated secondary Abs (1 hour at 37°C), and developed with p-nitrophenyl phosphate (1 hour at 37°C).
Immunocapture of C1q using DC-SIGN–conjugated beads
Human serum (150 μL) was first pre-absorbed with immobilized neutravidin Ultralink resin beads to minimize nonspecific binding (6 hours at 4°C). The serum was then incubated with 5 μg of biotinylated DC-SIGN (overnight at 4°C) and added to the beads containing 0.5M NaCl (1 hour at 4°C). C1q was used as a positive control. After incubation, the beads were washed 5 times with PBS containing 0.1% SDS, 0.5M NaCl, and 0.05% Tween-20 and the bound proteins were eluted with Laemmli sample buffer under reducing conditions and analyzed by Western blotting.
Western blotting
Proteins were separated by SDS-PAGE using a 12% gel, transferred to a nitrocellulose membrane, blocked in 5% milk (1 hour at room temperature [RT]), and incubated with primary Abs (overnight at 4°C). The membrane was washed in TBS containing 0.05% Tween and incubated with HRP-conjugated secondary Abs (1 hour at RT). The blot was developed using SuperSignal West Pico-ECL detection reagent according to the manufacturer's specifications and exposed to Kodak Scientific Imaging X-OMAT film.
SPR
For surface plasmon resonance (SPR) analysis, DC-SIGN (5 μg/mL) was immobilized on a BIAcore CM5 sensor chip at an immobilization level of 550 RU. C1q/gC1q was injected for 2 minutes at various concentrations (0.035-0.7μM for C1q and 0.156-1.25μM for gC1q). The complexes formed were allowed to dissociate by washing the surfaces with 10mM acetate buffer (pH 5.5) for 3 minutes. The DC-SIGN–bound surface was regenerated by injection of 10mM glycine (pH 2.0). All binding affinities were calculated using a 1:1 Langmuir fitting-based curve and BIAcore 3000 BIAevaluation software with an average value determined over the range of concentrations measured.
For assessing gC1qR–DC-SIGN binding, gC1qR was immobilized on a CM5 chip (60 μg/mL in 10mM acetate, pH 4.5, for 2 minutes at a 10 μL/min flow rate) at an immobilization level of 350 RU. To remove the avidity effects due to the tetrameric nature of DC-SIGN, a DC-SIGN R8 recombinant protein construct that is a monomeric form of DC-SIGN,31 was passed over immobilized gC1qR at multiple concentrations (1-100μM) at a flow rate of 40 μL/min until equilibrium was reached. Binding affinity was determined by plotting response versus concentration.
Competition studies
For all competition studies, ELISA experiments were performed as described above with minor adjustments. For the IgG competition study, 5 μg/mL of C1q was preincubated with human IgG at a 1:10 molar ratio (15 minutes at 37°C) and added to DC-SIGN–coated wells (1 hour at 37°C). C1q alone was used as a positive control.
For the C1q-derived synthetic peptide studies, after blocking, the wells were incubated with increasing concentrations of C1q-A peptide 155-164 (0-80 μg/mL). After washing, biotinylated C1q was added (1 hour at 37°C) and the binding was detected using streptavidin-AP.
For the mannan competition studies, C1q (5 μg/mL) or the synthetic peptide RR (50 μg/mL) was captured on the plate, blocked, and incubated with 2 mg/mL of mannan (as a negative control), biotinylated DC-SIGN (as a positive control), or biotinylated DC-SIGN that was preincubated with 2 mg/mL of mannan (15 minutes at RT).
For the C1q/gC1qR competition study, 5 μg/mL of biotinylated gC1qR was mixed with increasing concentrations of C1q and added to DC-SIGN–coated wells (2 hours at 37°C). Biotinylated gC1qR alone was used as a positive control.
Immunofluorescent microscopy
Cell surface staining was performed as described previously.9 Briefly, cells were incubated with rat anti–DC-SIGN, rabbit anti-C1q, or rabbit anti-gC1qR (30 minutes at 4°C), followed by secondary Abs (anti–rat FITC + anti–rabbit PE for 30 minutes at 4°C) with 1% DAPI and then fixed in 1% formalin. The cells were applied to microscope slides, air dried, and cover slipped using ImmunoMount mounting solution. Between each step, 2 washes were performed using PBA.
In addition, human tonsils obtained from the National Cancer Institute were analyzed for DC-SIGN and C1q. Cryostat sections were acetone fixed (1 minute at 4°C), rinsed in PBS, and blocked with 1% goat serum/PBS. The slides were incubated with rat anti–human DC-SIGN and goat anti–human C1q (2 hours at RT). The slides were washed, incubated with FITC- and PE-conjugated secondary Abs and DAPI (1 hour at RT), washed again, and cover slipped using ImmunoMount mounting solution. The slides were analyzed using an Axiovert 200M digital deconvolution microscope (Carl Zeiss) fitted with a Chroma filter set. Images were captured with a Peltier-cooled charge-coupled AxioCam HRm camera (Carl Zeiss) at 68× oil magnification and analyzed with AxioVision Version 4.8 software.
Signaling assay
THP-1 cells were preincubated with or without anti–DC-SIGN Abs (AZND1 and MR-1; 20 minutes at 37°C) before the addition of C1q (20 μg/mL for 10 minutes at 37°C). Cells were lysed in cell lysis buffer (20mM Tris-HCl, pH 7.5, 150mM NaCl, 1mM EDTA, 1% Triton X-100, 2.5mM Na-pyrophosphate, 1mM β-glycerophosphate, 1mM Na3VO4, and 1 μg/mL of leupeptin) and assayed by Western blotting.
Statistical analysis
Student t tests were performed using statistical software (Microsoft Excel 2010). P = .05 was considered to be a significant difference (n values represent separate experiments performed using different donors).
Results
Extracellular expression of DC-SIGN during the monocyte-DC transition
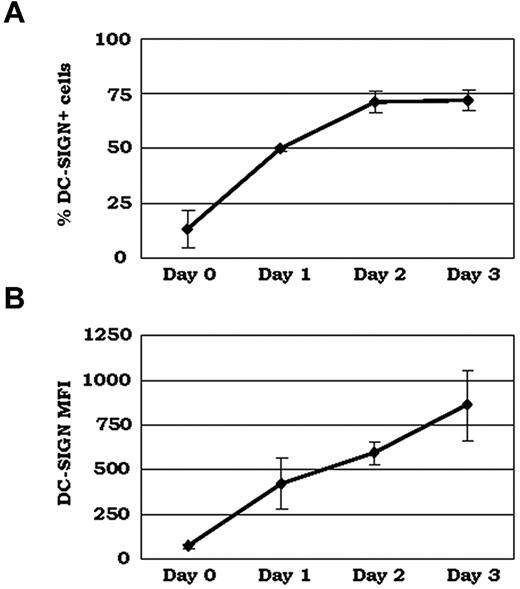
To investigate the hypothesis that DC-SIGN functions as a coreceptor for C1q, we first sought to confirm surface expression of DC-SIGN during the monocyte-to-DC transition. PBMCs were cultured with GM-CSF/IL-4 to generate monocyte-derived DCs and then analyzed by flow cytometry. DC-SIGN expression was observed on a small percentage of freshly isolated blood precursors that had not yet been exposed to DC growth factors (Figure 1A), confirming previous results.28,29 DC-SIGN expression increased markedly within 24 hours after GM-CSF/IL-4–induced DC differentiation (Figure 1). The percentage of cells expressing DC-SIGN on their surface increased until day 2, when 75%-80% of the cells had DC-SIGN (Figure 1A). Mean fluorescence intensities, indicating the relative abundance of DC-SIGN on each cell, continued to increase past day 2, and peaked after the cells had fully committed to the DC lineage (days 3-4; Figure 1B), as assessed by the marked up-regulation of DC maturation markers (CD86, HLA-DR, and CD11c; data not shown).
Surface expression of DC-SIGN increases as monocytes differentiate into iDCs. PBMCs were isolated by density gradient centrifugation and cultured for 3 days in the presence of DC growth factors. Cells were collected on each day and incubated with anti–DC-SIGN mAb, followed by incubation with a secondary Ab conjugated to Alexa Fluor 488 and analysis by flow cytometry. (A) Percentage of positive cells. (B) Mean fluorescence intensity (MFI; n = 3).
Surface expression of DC-SIGN increases as monocytes differentiate into iDCs. PBMCs were isolated by density gradient centrifugation and cultured for 3 days in the presence of DC growth factors. Cells were collected on each day and incubated with anti–DC-SIGN mAb, followed by incubation with a secondary Ab conjugated to Alexa Fluor 488 and analysis by flow cytometry. (A) Percentage of positive cells. (B) Mean fluorescence intensity (MFI; n = 3).
DC-SIGN binds to C1q
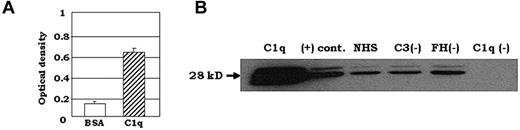
Previous work by Kang et al found that SIGN-R1, the murine homolog of DC-SIGN, bound directly to C1q.27 In the present study, we sought to determine whether DC-SIGN is also capable of binding to C1q. We examined direct binding of purified C1q on DC-SIGN–coated microtiter plate wells. Figure 2A shows the results of a representative antigen-capture ELISA in which the direct interaction of C1q with DC-SIGN was analyzed. These experiments showed strong binding of C1q to DC-SIGN, whereas negligible binding was observed when BSA was used instead of DC-SIGN (Figure 2A). Moreover, a similar interaction pattern was observed when ELISA was performed in the reverse configuration: capture of DC-SIGN with microplate-bound C1q (data not shown).
C1q binds to DC-SIGN. (A) C1q was added to DC-SIGN–coated plates and binding was detected using pAb anti-C1q Ab in an antigen-capture ELISA. One representative experiment is shown (n = 3). (B) Normal human serum (NHS; 150 μL) that was C3, factor H (FH), and C1q depleted was incubated overnight at 4°C with 2 μg of biotinylated DC-SIGN and added to neutravidin-coated resin beads for 2 hours at RT. Bead-bound proteins were visualized by Western blotting using pAb C1q, followed by HRP- conjugated secondary Ab. Soluble C1q protein was used instead of human serum as a positive control, shown as “(+) cont.” (n = 2).
C1q binds to DC-SIGN. (A) C1q was added to DC-SIGN–coated plates and binding was detected using pAb anti-C1q Ab in an antigen-capture ELISA. One representative experiment is shown (n = 3). (B) Normal human serum (NHS; 150 μL) that was C3, factor H (FH), and C1q depleted was incubated overnight at 4°C with 2 μg of biotinylated DC-SIGN and added to neutravidin-coated resin beads for 2 hours at RT. Bead-bound proteins were visualized by Western blotting using pAb C1q, followed by HRP- conjugated secondary Ab. Soluble C1q protein was used instead of human serum as a positive control, shown as “(+) cont.” (n = 2).
To confirm the interaction between C1q and DC-SIGN, we examined whether DC-SIGN–coated beads could capture C1q from solution. We immobilized biotinylated DC-SIGN on neutravidin-conjugated beads and mixed the beads with purified C1q. Elution of the bound protein followed by Western blotting showed that the DC-SIGN–coated beads captured soluble C1q (Figure 2B positive control). To elucidate the specificity of binding, we examined whether DC-SIGN–coated beads would pull down C1q from normal human serum, as well as from sera depleted of C3, factor H, and C1q. We mixed the sera with DC-SIGN–coated beads and detected the bound C1q by Western blotting. Whereas no C1q binding was detected when the C1q-depleted serum was incubated with DC-SIGN–coated beads, strong C1q binding was observed in both the factor H- and C3-depleted sera, and the intensity of the bands was similar to that of C1q captured from normal human serum (Figure 2B).
Plasmon resonance analysis
To explore the affinity of the C1q–DC-SIGN interaction, we monitored the binding of C1q and its globular region (gC1q) to a DC-SIGN–functionalized sensor chip by BIAcore SPR analysis using a BIAcore 3000 instrument. DC-SIGN (5 μg/mL in 10mM acetate, pH 5.5, for 2 minutes at a 10 μL/min flow rate) was coupled to a BIAcore CM5 dextran chip and various concentrations of C1q or gC1q were passed over the immobilized DC-SIGN at a flow rate of 20 μL/min. Both C1q and gC1q bound DC-SIGN in a dose-dependent manner, with a KD of 1.87 × 10−7M and 9.6 × 10−6M, respectively (Table 1 and supplemental Figure 1A-B, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The KD for the C1q-gC1qR interaction has been reported to be 2.4 × 10−7M,14 suggesting that C1q binds to gC1qR and DC-SIGN with similar affinities. In contrast, the control protein failed to demonstrate binding to DC-SIGN.
The results of these experiments show that C1q binds to DC-SIGN specifically, and suggest that it likely binds DC-SIGN through its gC1q region.
DC-SIGN binds to C1q at the IgG-binding site
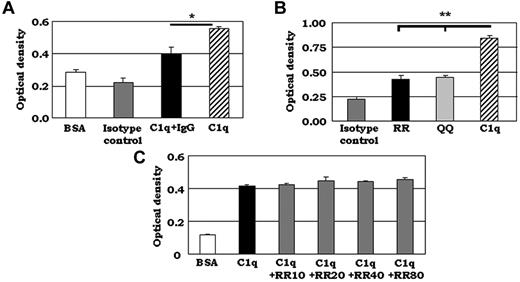
We also sought to identify possible DC-SIGN–binding sites on C1q. Because our SPR experiments had indicated that the gC1q region bound to DC-SIGN, we focused on this region. As reported previously, both gC1qR and IgG bound to the A-chain of gC1q.32 Therefore, we first examined whether DC-SIGN also bound to C1q at the same site. We assessed the effect of IgG on the C1q-DC-SIGN interaction by preincubating C1q with human IgG in a 1:10 molar ratio before adding it to DC-SIGN–coated microtiter wells. Whereas untreated C1q (a positive control) showed strong binding to DC-SIGN, preincubation with IgG resulted in a significantly reduced C1q–DC-SIGN interaction (Figure 3A). These results suggest that the DC-SIGN–binding site is either the same as or overlaps with the IgG-binding site on C1q-A.
DC-SIGN binds to C1q at the IgG-binding site. (A) IgG reduced the binding of C1q to DC-SIGN significantly. ELISA experiments were performed using C1q premixed with human IgG in a 1:10 molar ratio for 15 minutes at 37°C. C1q/IgG was added to DC-SIGN– or BSA-coated plates and binding was detected using C1q pAb. One representative experiment is shown (n = 3). *P < .05. (B) Synthetic peptide corresponding to C1q-A chain residues 155-164 bound to DC-SIGN, and the Arg residues (162-163) were not required for binding. ELISA binding studies were performed using 2 synthetic peptides, the C1q-A-chain peptide containing 2 adjacent Arg residues (RR) and another peptide that had 2 glutamines (QQ) substituting for the 2 Arg residues at positions 162-163 (n = 3). *P < .05. (C) Competition of a synthetic peptide corresponding to C1q-A 155-164 did not diminish binding of C1q to DC-SIGN. Competition ELISA experiments were performed using purified, soluble C1q and the synthetic C1q peptide RR. Biotinylated C1q and increasing concentrations of RR (0-80 μg/mL) were added to DC-SIGN–coated plates and binding was detected using anti-C1q oAb. One representative experiment is shown (n = 3).
DC-SIGN binds to C1q at the IgG-binding site. (A) IgG reduced the binding of C1q to DC-SIGN significantly. ELISA experiments were performed using C1q premixed with human IgG in a 1:10 molar ratio for 15 minutes at 37°C. C1q/IgG was added to DC-SIGN– or BSA-coated plates and binding was detected using C1q pAb. One representative experiment is shown (n = 3). *P < .05. (B) Synthetic peptide corresponding to C1q-A chain residues 155-164 bound to DC-SIGN, and the Arg residues (162-163) were not required for binding. ELISA binding studies were performed using 2 synthetic peptides, the C1q-A-chain peptide containing 2 adjacent Arg residues (RR) and another peptide that had 2 glutamines (QQ) substituting for the 2 Arg residues at positions 162-163 (n = 3). *P < .05. (C) Competition of a synthetic peptide corresponding to C1q-A 155-164 did not diminish binding of C1q to DC-SIGN. Competition ELISA experiments were performed using purified, soluble C1q and the synthetic C1q peptide RR. Biotinylated C1q and increasing concentrations of RR (0-80 μg/mL) were added to DC-SIGN–coated plates and binding was detected using anti-C1q oAb. One representative experiment is shown (n = 3).
To further explore the extent to which the DC-SIGN- and IgG-binding sites might overlap, we focused on the Arg residues at positions 162-163 of C1q-A, which are involved in IgG and gC1qR binding.32 We generated 2 synthetic peptides: one peptide (RR) corresponded to C1q-A chain residues 155-164 (SSSRGQVRRS) and the other (QQ) was identical except the Arg residues at positions 162-163 were substituted by Gln (SSSRGQVQQS). Interestingly, both peptides bound to DC-SIGN, however, to a significantly lesser extent than intact C1q (Figure 3B). The level of binding between C1q and DC-SIGN could not be achieved even when the concentration of the synthetic peptides was increased to 3-fold higher than that of C1q (data not shown). These results indicate that DC-SIGN binds to C1q-A at the gC1qR/IgG–binding site, but that Arg residues are not required for binding.
To compare the relative binding ability of C1q and the Arg-containing synthetic peptide RR, competition binding assays of biotinylated C1q with various concentrations of RR were performed. Binding of C1q to DC-SIGN was not blocked by coincubation with the RR peptide (Figure 3C), suggesting that residues 155-164 do not span the entire DC-SIGN–binding site or that there are alternative binding sites present.
The C1q–DC-SIGN interaction is Ca2+ dependent
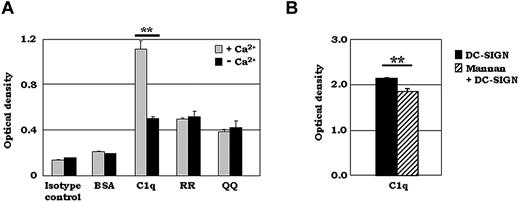
Many ligands that bind to DC-SIGN are heavily glycosylated (eg, ICAM 2/3) and the glycosylation-specific nature of these interactions is Ca2+ dependent.33 Because C1q is glycosylated, we hypothesized that Ca2+ may also be necessary for its recognition by DC-SIGN. To test this hypothesis, we compared the binding of C1q and synthetic C1q peptides (RR and QQ) to DC-SIGN in the presence and absence of EDTA. As expected, the binding of the unglycosylated synthetic C1q-A peptides was not affected by the presence of 10mM EDTA (Figure 4A). In contrast, C1q binding, which was markedly higher than that of either C1q-A peptide in the presence of Ca2+, was reduced significantly in the absence of Ca2+ (Figure 4A). Binding of the unglycosylated synthetic peptides suggests that amino acid residues on both molecules also contribute to the binding.
The C1q–DC-SIGN interaction is Ca2+ dependent and reduced by mannan. (A) ELISA experiments were performed using purified, soluble C1q or various synthetic C1q-A peptides (aa 155-164 for RR, aa 155-164 for RR at 162/163 changed to QQ). Then, 2.5 μg/mL of biotinylated DC-SIGN was added to the C1q- or synthetic peptide–coated plates in the presence or absence of 10mM EDTA and binding was detected using streptavidin-AP. One representative experiment is shown (n = 3). (B) Mannan reduced the binding of C1q to DC-SIGN significantly. ELISA experiments were performed using 2.5 μg/mL of biotinylated DC-SIGN premixed with 2 mg/mL of mannan for 15 minutes at RT. DC-SIGN/mannan was added to C1q- or BSA-coated plates and binding was detected using streptavidin-AP. One representative experiment is shown (n = 3). **P < .01.
The C1q–DC-SIGN interaction is Ca2+ dependent and reduced by mannan. (A) ELISA experiments were performed using purified, soluble C1q or various synthetic C1q-A peptides (aa 155-164 for RR, aa 155-164 for RR at 162/163 changed to QQ). Then, 2.5 μg/mL of biotinylated DC-SIGN was added to the C1q- or synthetic peptide–coated plates in the presence or absence of 10mM EDTA and binding was detected using streptavidin-AP. One representative experiment is shown (n = 3). (B) Mannan reduced the binding of C1q to DC-SIGN significantly. ELISA experiments were performed using 2.5 μg/mL of biotinylated DC-SIGN premixed with 2 mg/mL of mannan for 15 minutes at RT. DC-SIGN/mannan was added to C1q- or BSA-coated plates and binding was detected using streptavidin-AP. One representative experiment is shown (n = 3). **P < .01.
DC-SIGN has increased affinity for high mannose residues, and its principal Ca2+-binding pocket also accommodates mannose moieties. Because the carbohydrate residues on C1q contain mannose,34 we next determined the carbohydrate specificity of the C1q–DC-SIGN interaction by inhibition assays using mannan, a mannose polymer. As shown in Figure 4B, the addition of 2 mg/mL of mannan reduced the binding activity of C1q to DC-SIGN significantly, suggesting the involvement of mannose in C1q recognition.
These data suggest that the C1q–DC-SIGN interaction depends on the presence of Ca2+ and that the affinity of binding may be highly increased by the specific carbohydrate moieties present on C1q.
DC-SIGN colocalizes with C1q on the surface of DC blood precursors in vivo and in vitro
We have reported previously the presence of C1q on the surface of PBMCs and monocyte-derived iDCs.9 In the present study, we sought to determine whether endogenously expressed DC-SIGN interacts with C1q on the surface of these cells. We first verified the interaction by capturing C1q from the lysates of day 3 iDCs on microtiter plate wells coated with pAb anti–DC-SIGN, followed by detection for C1q. The results of a representative antigen-capture ELISA illustrated that C1q could be cocaptured with DC-SIGN, whereas an isotype control Ab showed negligible binding (Figure 5A). To authenticate the specificity of these results, we performed the ELISA in reverse, first capturing C1q and detecting for DC-SIGN, with similar results (data not shown). These data suggest that DC-SIGN either directly or indirectly partners with C1q on iDCs.
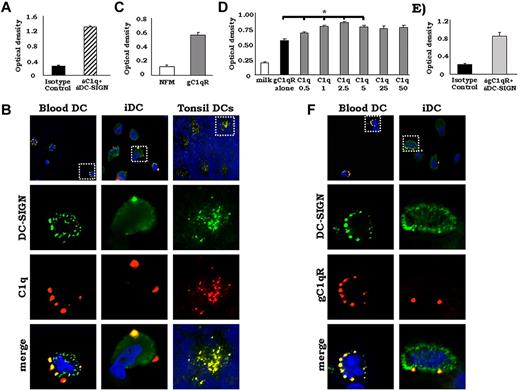
DC-SIGN colocalized with C1q and gC1qR on DCs. (A) DC-SIGN was cocaptured with C1q from iDC lysates. Whole-cell lysates (day 3) were added to microtiter plates coated with mAb DC-SIGN and the presence of C1q was detected using anti-C1q pAb. One representative experiment is shown (n = 3). (B) DC-SIGN colocalizes with C1q on the surface of iDCs in vitro, on DC precursors in blood and on iDCs in human tonsils in vivo. PBMCs, day 3 iDCs, and cryostat tonsil sections were analyzed for DC-SIGN and C1q. Cells and tonsil sections were incubated with rat anti–DC-SIGN and goat anti-C1q Abs or isotype controls, followed by FITC–anti–rat and PE-anti–goat secondary Abs and DAPI (blue). The slides were viewed using a Zeiss Axiovert 200M digital deconvolution microscope (63×; oil) and analyzed with AxioVision Version 4.8 software. Isotype controls showed little or no staining (data not shown; n = 6). (C) DC-SIGN binds to gC1qR. Antigen-capture ELISA experiments were performed using purified, soluble gC1qR. gC1qR was added to DC-SIGN–coated plates and binding was detected using anti-gC1qR pAb. One representative experiment is shown (n = 3). (D) C1q increased the binding of gC1qR to DC-SIGN significantly. ELISA experiments were performed using 5 μg/mL of biotinylated gC1qR premixed with increasing concentrations (0-50 μg/mL) of C1q. Biot-gC1qR/C1q was added to DC-SIGN– or 5% nonfat milk–coated plates and binding was detected using neutravidin-AP. One representative experiment is shown (n = 2). *P < .05. (E) DC-SIGN was cocaptured with gC1qR from iDC lysates. Antigen-capture ELISA experiments were performed using whole-cell DC lysates (day 3). DC lysates were added to microtiter plates coated with rat anti–human DC-SIGN and the presence of gC1qR was detected using anti-gC1qR pAb. One representative experiment is shown (n = 3). (F) DC-SIGN colocalizes with gC1qR on the surface of blood DC precursors and iDCs. PBMCs and day 3 iDCs were incubated with rat anti–DC-SIGN and rabbit anti-gC1qR Abs or isotype controls, followed by FITC-anti–rat and PE–anti–rabbit secondary Abs and DAPI (blue). The slides were viewed using a Zeiss Axiovert 200M digital deconvolution microscope (68×; oil) and analyzed with AxioVision Version 4.8 software. Isotype controls showed little or no staining (data not shown; n = 7).
DC-SIGN colocalized with C1q and gC1qR on DCs. (A) DC-SIGN was cocaptured with C1q from iDC lysates. Whole-cell lysates (day 3) were added to microtiter plates coated with mAb DC-SIGN and the presence of C1q was detected using anti-C1q pAb. One representative experiment is shown (n = 3). (B) DC-SIGN colocalizes with C1q on the surface of iDCs in vitro, on DC precursors in blood and on iDCs in human tonsils in vivo. PBMCs, day 3 iDCs, and cryostat tonsil sections were analyzed for DC-SIGN and C1q. Cells and tonsil sections were incubated with rat anti–DC-SIGN and goat anti-C1q Abs or isotype controls, followed by FITC–anti–rat and PE-anti–goat secondary Abs and DAPI (blue). The slides were viewed using a Zeiss Axiovert 200M digital deconvolution microscope (63×; oil) and analyzed with AxioVision Version 4.8 software. Isotype controls showed little or no staining (data not shown; n = 6). (C) DC-SIGN binds to gC1qR. Antigen-capture ELISA experiments were performed using purified, soluble gC1qR. gC1qR was added to DC-SIGN–coated plates and binding was detected using anti-gC1qR pAb. One representative experiment is shown (n = 3). (D) C1q increased the binding of gC1qR to DC-SIGN significantly. ELISA experiments were performed using 5 μg/mL of biotinylated gC1qR premixed with increasing concentrations (0-50 μg/mL) of C1q. Biot-gC1qR/C1q was added to DC-SIGN– or 5% nonfat milk–coated plates and binding was detected using neutravidin-AP. One representative experiment is shown (n = 2). *P < .05. (E) DC-SIGN was cocaptured with gC1qR from iDC lysates. Antigen-capture ELISA experiments were performed using whole-cell DC lysates (day 3). DC lysates were added to microtiter plates coated with rat anti–human DC-SIGN and the presence of gC1qR was detected using anti-gC1qR pAb. One representative experiment is shown (n = 3). (F) DC-SIGN colocalizes with gC1qR on the surface of blood DC precursors and iDCs. PBMCs and day 3 iDCs were incubated with rat anti–DC-SIGN and rabbit anti-gC1qR Abs or isotype controls, followed by FITC-anti–rat and PE–anti–rabbit secondary Abs and DAPI (blue). The slides were viewed using a Zeiss Axiovert 200M digital deconvolution microscope (68×; oil) and analyzed with AxioVision Version 4.8 software. Isotype controls showed little or no staining (data not shown; n = 7).
We also investigated whether DC-SIGN and C1q colocalize on the surface of DC blood precursors and iDCs using immunofluorescent microscopy. Qualitative analysis of colocalization showed that nearly all C1q colocalized with DC-SIGN on the surface of DC precursors freshly isolated from the blood (Figure 5B blood DC). Colocalization was still evident on day 3 iDCs; however, some patches of C1q were also detectable as separate clusters (Figure 5B iDC). Furthermore, the distribution pattern of both C1q and DC-SIGN on the plasma membrane changed during cell differentiation. Microscopic analyses revealed a punctate pattern of expression for both molecules on the surface of freshly isolated blood precursors evenly distributed over the entire cellular membrane (Figure 5B blood DC). By day 3, when the majority of observed cells were DC like in phenotype, C1q expression was confined to large patches, whereas DC-SIGN was seen throughout the cell surface and distributed in a more even pattern (Figure 5B iDC).
We next investigated whether the colocalization of DC-SIGN and C1q also exists in vivo on iDCs. Using a mAb against DC-SIGN, we identified DC-SIGN+ iDCs localized to the densely cellular paracortical areas of the human tonsil, which, in addition to a network of interdigitating DCs, contain lymphoid cells predominantly of the T-cell type (Figure 5B tonsil DC). Double-staining experiments confirmed that DC-SIGN and C1q colocalized on the surface of these cells (Figure 5B tonsil DC).
DC-SIGN interacts with the C1q receptor gC1qR
Because DC-SIGN bound to gC1q (Table 1), we next sought to determine whether DC-SIGN was also capable of binding to gC1qR. In antigen-capture experiments, DC-SIGN coated on microtiter plate wells was able to capture soluble gC1qR (Figure 5C). Furthermore, binding of gC1qR to a DC-SIGN–functionalized sensor chip by SPR showed that gC1qR bound to DC-SIGN with a KD of 40 × 10−6M (Table 1 and supplemental Figure 1C). To determine whether C1q and gC1qR compete for the same binding site on DC-SIGN, competition assays using biotinylated-gC1qR with various concentrations of C1q were performed. Interestingly, binding of gC1qR to DC-SIGN was enhanced significantly by coincubation with C1q (Figure 5D), suggesting that C1q binds multiple units of gC1qR and thus increases its binding avidity to DC-SIGN. When the reverse assay was performed (biotinylated C1q with increasing concentrations of gC1qR added to DC-SIGN–coated plates until binding of biot-C1q was detected), no significant change of C1q binding was observed (data not shown). These data suggest that C1q and gC1qR do not compete for the same binding site.
We also sought to determine whether DC-SIGN associates with gC1qR on day 3 iDCs. Using whole-cell lysates generated from day 3 cultured iDCs, we captured DC-SIGN from the lysates on microtiter plate wells using pAbs, followed by detection for gC1qR. GC1qR could be cocaptured with DC-SIGN, whereas an isotype control Ab showed very little binding (Figure 5E).
Immunofluorescent microscopy revealed that DC-SIGN colocalized with gC1qR on the surface of freshly isolated DC precursors (Figure 5F blood DC). On day 3 iDCs, approximately 50% of DC-SIGN and gC1qR was still observed to colocalize on each cell (Figure 5F iDC); however, some patches of gC1qR localized separately from DC-SIGN, suggesting that this association was not ubiquitous. Similarly to C1q and DC-SIGN, the distribution pattern of gC1qR on the plasma membrane changed during cell differentiation. Whereas gC1qR exhibited a punctate pattern of expression on fresh DC precursors, on day 3 iDCs, gC1qR was localized to specific regions (Figure 5F iDC). These results show that, similar to C1q and DC-SIGN, the surface distribution of gC1qR varies with DC differentiation and that gC1qR and DC-SIGN colocalize on the surface of DCs.
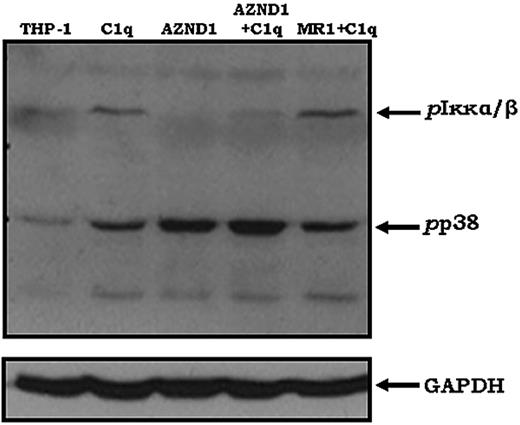
C1q–DC-SIGN interaction activates NF-κB
The binding of C1q to DCs induced the expression of specific pro- and anti-inflammatory cytokines (data not shown); therefore, we investigated the phosphorylation levels of Iκκα/β, members of the IκB kinase complex that phosphorylate IκB, thereby activating NF-κB. A DC-SIGN–inhibiting Ab (AZND1) abrogated the C1q-induced phosphorylation of Iκκα/β completely, as assessed by Western blotting, whereas another DC-SIGN–specific Ab (MR-1) had no effect (Figure 6). Because the activation of p38 MAPK can also influence gene transcription directly, resulting in cell differentiation and cytokine production, we analyzed p38 phosphorylation. Treatment with C1q increased phosphorylated p38 MAPK levels compared with untreated THP-1 cells. Surprisingly, the AZND-1 Ab alone also increased p38 phosphorylation. The addition of C1q after preincubating the cells with AZND-1 showed a synergistic effect, increasing phosphorylated p38 levels even further (Figure 6). The MR-1 Ab had no effect on C1q-mediated p38 phosphorylation. These results suggest that C1q can activate intracellular signaling pathways through DC-SIGN specifically.
C1q-mediated activation of NF-κB requires DC-SIGN. Phosphorylation of Iκκα/β in the DC-SIGN–expressing human THP-1 cell line was determined after treatment with Abs to DC-SIGN (AZND1 and MR-1; 20 μg/mL at 37°C for 20 minutes) and/or C1q (at 37°C for 10 minutes). After cell lysis, phosphorylated Iκκα/β and p38 were detected using specific mAbs. The blot was then stripped and probed for GAPDH levels as a control for protein loading (n = 2).
C1q-mediated activation of NF-κB requires DC-SIGN. Phosphorylation of Iκκα/β in the DC-SIGN–expressing human THP-1 cell line was determined after treatment with Abs to DC-SIGN (AZND1 and MR-1; 20 μg/mL at 37°C for 20 minutes) and/or C1q (at 37°C for 10 minutes). After cell lysis, phosphorylated Iκκα/β and p38 were detected using specific mAbs. The blot was then stripped and probed for GAPDH levels as a control for protein loading (n = 2).
Discussion
We showed previously that C1q regulates the monocyte-to-DC transition by acting as a molecular switch that skews DC differentiation toward specific iDC subsets.9 The present study was undertaken to gain insight into the mechanism by which C1q fulfills its regulatory function.
The data presented herein reveal several novel observations. First, C1q binds to DC-SIGN primarily via its gC1q. Second, binding of C1q is reduced significantly by preincubation with IgG, suggesting that DC-SIGN and IgG compete for an overlapping site on C1q. Third, the C1q–DC-SIGN interaction is reduced in the absence of Ca2+ and by preincubation of DC-SIGN with mannan. This suggests that C1q binds to DC-SIGN at its principal Ca2+-binding pocket, which has increased affinity for high mannose residues, and that carbohydrate moieties on C1q contribute to the C1q–DC-SIGN interaction. However, because mannan, even in excess of 1000-fold, could not abrogate C1q binding to DC-SIGN, protein-protein interactions may also play a role in forming the bond. Fourth, C1q and gC1qR associate with DC-SIGN on the surface of iDCs and blood DC precursors. Because gC1qR lacks a consensus motif for a transmembrane segment, it is postulated to trigger downstream signaling by forging a docking/signaling partnership with transmembrane proteins. This partnership is dictated by the cell type and the biologic response to be induced. Although the signaling mechanism has yet to be determined, the finding that C1q, DC-SIGN, and gC1qR form a complex on the DC surface suggests a role of C1q/gC1qR in the regulation of DC differentiation and function through DC-SIGN–mediated induction of cell-signaling pathways.
The results of the present study show that DC-SIGN binds to monomeric gC1q directly and specifically, albeit with slightly lower affinity than to intact C1q. Its affinity would be expected to increase within the “normal” structure of hexameric C1q. Whereas the data indicate that C1q binds to DC-SIGN via its gC1q, we cannot rule out potential binding sites on the cC1q domain. Our SPR experiments showed that monomeric DC-SIGN binds to C1q and gC1qR with a KD of 1.87 × 10−7M and 40 × 10−6M, respectively. Frison et al showed that the binding specificity of DC-SIGN for multiple repetitive units on host molecules is amplified when DC-SIGN is tetramerized.35 Therefore, we hypothesize that the hexameric C1q and the trimeric gC1qR bind to tetramerized DC-SIGN with very high affinity.
Although more work is needed, we have begun to unravel the biologic significance of the C1q–DC-SIGN interaction. Our results show that AZND1, a DC-SIGN–inhibiting Ab, could completely abrogate the C1q-induced phosphorylation of Iκκα/β in the DC-SIGN–expressing THP-1 cells, whereas another DC-SIGN–specific Ab (MR-1) had no effect. These results suggest that C1q can activate the NF-κB pathway via DC-SIGN, specifically through a binding site that overlaps with that of the AZND1 Ab. Furthermore, we observed a synergistic increase in the phosphorylation of p38 MAPK when THP-1 cells were treated with a combination of C1q and AZND-1. These results suggest that p38 activation may play a role in the C1q-induced regulation of DC differentiation.
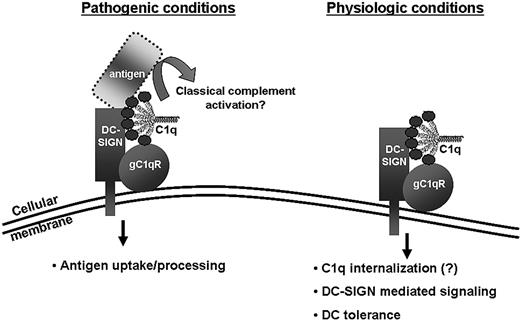
We have highlighted a hypothetical model based on the data presented in these studies (Figure 7). Under physiologic conditions, “empty” C1q may bind to the gC1qR–DC-SIGN complex and initiate DC-SIGN–mediated cellular signaling and DC tolerance. It remains to be determined whether, on binding to DC-SIGN, C1q is taken up by these cells, because the di-leucin motifs on the cytoplasmic tail of DC-SIGN have been shown to induce rapid internalization.36 Such uptake of Ag-free C1q may reflect a regulatory mechanism that sustains innate immune functions and may act through a DC-SIGN–mediated signaling mechanism. Under pathogenic conditions, antigen-complexed C1q may bind to gC1qR and facilitate antigen uptake via DC-SIGN. Concurrently, the C1q–DC-SIGN complex may provide an initiation site for the classic complement pathway in the presence of a pathogenic surface. Such a scenario was proposed by Kang et al27 based on their studies of C1q-SIGN-R1 interaction. In their hypothetical model, SIGN-R1, which binds polysaccharides and pneumococci on macrophages, may replace the Ig usually used in the classic pathway to bind C1q. The C1q–SIGN-R1 complex may then assemble a C3 convertase and perpetuate the complement cascade. In this setting, C1q would function as a sentinel of danger, probably reflecting its earliest ability to obtain extracellular “danger” Ag for complement initiation and reentry into DCs for processing. We believe that signaling and immunologic outcome dictated by DC differentiation depends on whether the cargo carries a “danger” element such as PAMPs, “modified self-antigens,” or damage-associated molecular patterns. The conformational change in C1q/gC1qR on binding to pathogen may influence the binding affinity to DC-SIGN and thus determine the signaling outcome.
Hypothetical model for the gC1qR/C1q–DC-SIGN complex under pathologic and physiologic conditions.
Hypothetical model for the gC1qR/C1q–DC-SIGN complex under pathologic and physiologic conditions.
Binding of C1q was reduced significantly by preincubation with IgG, suggesting that DC-SIGN binds to C1q at the IgG-binding site. However, the Arg residues (162-163) that are pivotal in the interaction between IgG and the C1q-A chain were not required for DC-SIGN binding, and the binding was also not blocked by coincubation with a peptide corresponding to residues 155-164 on the C1q-A chain, which was identified previously as the binding site for both IgG and gC1qR. This suggests that whereas the binding sites may overlap, the DC-SIGN–binding site spans a larger segment on C1q. Multiple DC-SIGN–binding sites might be present on the C1q molecule, possibly corresponding to other IgG-C1q interaction sites. These sites have been identified to contain Arg residues pivotal in the IgG-C1q bond (ArgA162, ArgB114, ArgB129, ArgB163, and ArgC156), pinpointing the importance of all 3 C1q chains in binding to IgG and suggesting that all 3 C1q chains support IgG binding by providing residues that form a cluster in space.32
Our previous results using C1q domain–specific Abs9 suggested that the C1q on the surface of DC precursors is bound via the globular heads, presumably to gC1qR. In the present study, we have shown that C1q and gC1qR associate with DC-SIGN on blood DC precursors and iDCs. In both previous studies37,38 and our unpublished data, both DC-SIGN37 and gC1qR38 have been shown to colocalize to lipid rafts, which may further enable the receptors to engage in complex formation and signal transduction and increase their binding capacity through clustering. Interestingly, our data indicate that whereas nearly all C1q and gC1qR colocalized with DC-SIGN on blood DC precursors, only partial colocalization could be detected on iDCs. The functional significance of individually localized C1q and gC1qR on these cells may represent distinct roles for the molecules during this stage of DC differentiation.
Strikingly, gC1qR and DC-SIGN share the ability to bind several viral pathogens, including HIV,33,39 hepatitis C virus (HCV),40,41 adenovirus,42,43 and herpes simplex virus.44,45 Common in these viral infections is their ability to evade the immune system and cause chronic infections. HCV, which is highly efficient in establishing a persistent infection, curbs the immune system effectively through suppressing the activation and induction of proinflammatory responses by APCs and in turn inducing T-cell impairment. HCV core protein interaction with gC1qR on DCs inhibits TLR-induced IL-12 production and reduces IFN-γ secretion by allogeneic CD4+ T lymphocytes.46 Conversely, DC-SIGN binds to the HCV envelope glycoprotein E2 with high affinity and targets the virus to the liver and the infection of resident DCs.40 Although a collaborative role remains to be identified, the gC1qR–DC-SIGN system may work together to assist viral entry and the subsequent alteration of inflammatory responses by DCs during an HCV infection. During HIV-1 infection, DC-SIGN is thought to capture the virus at mucosal sites of entry, facilitating transport to lymphoid tissues, where DC-SIGN transmits HIV to T cells efficiently.33 This may work in collaboration with gC1qR, which has been shown to bind directly to HIV-gp41 and trigger the depletion of uninfected CD4+ T cells during infection.47 Through association on the cell surface, DC-SIGN and gC1qR/C1q may form a complex that is important in pathogen entry and concomitant immunosuppression by the regulation of immunity and the induction of tolerance. This novel association could be involved in the pathogenesis of certain chronic diseases through exploitation of the natural function of these receptors and the evasion of the adaptive immune response. Further investigation of the relationship between DC-SIGN and C1q/gC1qR may help us to better understand the process of DC differentiation (the mechanisms by which it is regulated and the signaling pathways involved) and may also reveal possible mechanisms of modulating DC function in diseases such as HIV-AIDS.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Daniel Mockler for explaining human tonsil anatomy and histology and Gregory Snyder for producing the DC-SIGN R8 protein.
This work was supported in part by grants from the National Institutes of Health (R01 AI 060866 and R01 AI 084178).
National Institutes of Health
Authorship
Contribution: K.K.H. designed and performed the experiments, analyzed the data, and wrote the manuscript; A.V. designed and performed the experiments and wrote the manuscript; U.V., R.V., M.G.J., and Y.J. performed the experiments; E.I.B.P. contributed vital ideas and reagents; and B.G. designed the experiments and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Berhane Ghebrehiwet, Stony Brook University School of Medicine, Health Sciences Center, T-16, Rm 040, Stony Brook, NY 11794-8161; e-mail: berhane.ghebrehiwet@stonybrook.edu.