Abstract
The primary consequence of positive selection is to render thymocytes responsive to cytokines and chemokines expressed in the thymic medulla. In the present study, our main objective was to discover which cytokines could support the differentiation of positively selected thymocytes. To this end, we have developed an in vitro model suitable for high-throughput analyses of positive selection and CD8 T-cell differentiation. The model involves coculture of TCRhiCD5intCD69− double-positive (DP) thymocytes with peptide-pulsed OP9 cells and γc-cytokines. We report that IL-4, IL-7, and IL-21 have nonredundant effects on positively selected DP thymocytes. IL-7 signaling phosphorylates STAT5 and ERK; induces Foxo1, Klf2, and S1pr1; and supports the differentiation of classic CD8 T cells. IL-4 activates STAT6 and ERK and supports the differentiation of CD8intPD-L1hiCD44hiEOMES+ innate CD8 T cells. IL-21 is produced by thymic epithelial cells and the IL-21 receptor-α is strongly induced on DP thymocytes undergoing positive selection. IL-21 signaling phosphorylates STAT3 and STAT5, but not ERK, and does not support CD8 T-cell differentiation. However, IL-21 has a unique ability to up-regulate BCL-6, expand DP thymocytes undergoing positive selection, and increase the production of mature T cells. Our data suggest that injection of recombinant IL-21 might enhance thymic output in subjects with age- or disease-related thymic atrophy.
Key Points
We present a novel system for high-throughput analyses of thymocyte selection and differentiation.
We report that IL-4, IL-7, and IL-21 have nonredundant effects on positively selected thymocytes.
Introduction
The T-cell repertoire is molded by elimination of the double-positive (DP) thymocytes (approximately 95%) the TCR of which interacts with insufficient or excessive affinity toward self-peptide/MHC (pMHC) complexes. The role of negative selection, the deletion of strongly self-reactive thymocytes, is straightforward: to prevent autoimmunity.1,2 The consequences of positive selection, the rescue of weakly self-reactive thymocytes from cell death, are less obvious and more complex. Recent studies in Bcl-2 transgenic Quad-deficient mice (B2m−/−H-2-Ab1−/−Cd4−/−Cd8a−/−) have shown that positive selection ensures the MHC restriction of the αβTCR repertoire; in the absence of positive selection, TCRs would resemble Abs and recognize MHC-independent ligands.3 Moreover, during positive selection, persistent TCR signaling drives DP2 thymocytes (TCRintCD5hi) to differentiate into CD4 T cells, whereas on cessation of TCR signaling, DP thymocytes become responsive to γc-cytokines, adopt a DP3 phenotype (TCRhiCD5int), up-regulate CCR7, and differentiate into CD8 T cells.4,5 For CD8 T cells, the key roles of TCR signals eliciting positive selection are therefore to convert DP thymocytes into cytokine-responsive thymocytes and to induce their migration toward the medulla.6,7 DP thymocytes signaled via TCRs to undergo positive selection must absolutely receive γc cytokine signals to differentiate into CD8 single-positive (SP) T cells.
Which pMHC complexes and γc-cytokines are normally instrumental in positive selection and intrathymic differentiation of CD8 T cells? Few pMHC complexes can support positive selection, but each “successful” one can select a diverse CD8 T-cell repertoire.8 Positive selection of an immunocompetent and self-protective repertoire of CD8 T cells depends on peptides generated by the thymoproteasome, which is present only in cortical thymic epithelial cells (TECs).9 Nonetheless, the precise biochemical structure of cortical TEC pMHC complexes that support positive selection is basically unknown and awaits characterization by high-throughput mass spectrometry analyses.10,11 Cytokines that signal through the common cytokine receptor γ chain include IL-2, IL-4, IL-7, IL-9, IL-15, and IL-21. The receptor for each γc cytokine activates JAK1 and JAK3, and the signals are transduced by STAT proteins.12 Given the profound T-cell deficit in the thymus and periphery of IL-7Rα–deficient humans and mice, IL-7 is undoubtedly the dominant γc cytokine for T-cell development.13,14 However, the contingency of T-cell production on IL-7 provision can be explained by the fact that IL-7 is essential for the expansion and survival of thymocyte precursors (before β-selection) and of naive SP T cells (in the periphery). We have no conclusive evidence that IL-7 has a nonredundant role in the intrathymic maturation of CD8 T cells after initiation of positive selection by TCR signals. In fact, the generation of a specific subset of “innate polyclonal” CD8 thymocytes was recently found to be dependent on IL-4 provision in addition to TCR signals.15,16
The fetal thymic organ culture model and its variants have been invaluable for dissecting the mechanisms of positive selection in vitro.17 However, these systems are hampered by several technical intricacies and cannot be used for high-throughput studies required to gain further insights into the role of specific peptides and cytokines in positive selection and T-cell differentiation.17 We have developed a novel in vitro model that is suitable for analysis of positive selection and T-cell differentiation and is based on coculture of CD69− DP3 thymocytes with peptide-pulsed OP9 stromal cells and γc-cytokines. This system is simple, flexible, and enables high-throughput analyses of various peptide-cytokine combinations. Using this model, in the present study, we performed comprehensive analyses of the effects of γc-cytokines and report that, after positive selection, recombinant IL-4 (rIL-4), rIL-7, and rIL-21 have nonredundant effects on the expansion of DP thymocytes and CD8 T-cell differentiation.
Methods
Mice
OT-I [B6.129S7-Rag1tm1Mom Tg(TcraTcrb)1100Mjb N9+N1] mice were from Taconic Farms. B2m−/− (B6.129P2-B2mtm1Unc/J), and C57BL/6 mice were from The Jackson Laboratory. All mice were housed under specific pathogen-free conditions and all experimental protocols were approved by the animal care and use committee of the University of Montréal.
Abs, cytokines, and reagents
Recombinant cytokines were from PeproTech. Abs used in flow cytometry and the Cytofix/Cytoperm kit for intracellular staining were from BD Pharmingen. The anti-PLZF Ab was from Abcam. Peptides were synthesized by GenScript and the SIINFEKL-tetramers kit was obtained from IBA. TL tetramers were a generous gift from Dr Hilde Cheroutre (La Jolla Institute for Allergy & Immunology, La Jolla, CA). CFSE (Invitrogen) and Dynabeads CD3/CD28 T Cell Expander kits (Life Technologies) were used according to the manufacturer's instructions.
Enrichment of primary TECs
Positive selection of OT-I DP thymocytes by peptide-pulsed OP9-GFP cells
OP9-GFP cells were kindly provided by Dr J. C. Zúñiga-Pflücker (University of Toronto, Sunnybrook and Women's Research Institute, Toronto, ON). Endogenous peptides presented by MHC I on the surface of OP9-GFP cells were eluted with acid treatment as described previously.11,20 For peptide loading, 105 OP9-GFP cells were resuspended in 1 mL complete α-MEM media containing a range of peptide concentration (0.01-100 μg/mL). For positive selection studies, 1 μg/mL of the selected peptide was used to load OP9-GFP cells for 4 hours at 37°C in 24-well plates. Cells were then gently washed and cocultured with 5 × 104 sorted DP thymocytes (unless specified otherwise) in complete culture medium (RPMI, 10% FBS, 5mM l-glutamine, 10mM sodium pyruvate, and 25mM β-mercaptoethanol) supplemented with the selected recombinant cytokine at the appropriate concentration (2 ng/mL for rIL-4, rIL-7, and rIL-13 and 10 ng/mL for rIL-21). Three days later, cells were analyzed for the generation of SP CD8 T cells by flow cytometry.
Western blots
Protein detection by Western blot was performed as described previously.19
CFSE dilution assays
Sorted thymocytes were labeled with 5 μg/mL of CFSE for 10 minutes at 37°C and washed once with complete medium and then 3 times with PBS. To assess the proliferation induced after the addition of rIL-21 (10 ng/mL), CFSE-labeled cells were cultured for 48 hours and CFSE staining intensity was analyzed by flow cytometry.
CD8 T-cell stimulation in vitro
After differentiation in vitro, 5 × 104 sorted CD8 T cells were cultured for 3 days at 37°C in medium supplemented with mouse rIL-2 (30 U/mL) and CD3/CD28 Dynabeads at a bead-to-cell ratio of 1:1.
Gene-expression analysis by qPCR
Thymocytes were sorted directly in 900 μL TRIzol (106 cells per tube), cells were lysed and the RNA was extracted in TRIzol reagent. A further RNA purification step was carried out using the RNA extraction kit (QIAGEN). Reverse transcription was carried out using a high-capacity cDNA reverse transcription kit and quantitative PCR (qPCR) was performed with a 7900HT Fast Real-Time PCR system at the genomic core facility of the Institute for Research in Immunology and Cancer. Target gene values were normalized to endogenous control Gapdh.
Intracellular staining
For analysis of phosphorylated AKT (pAKT), pERK, pZAP70, pSTAT3, pSTAT5, and pSTAT6, sorted DP thymocytes were cocultured with OP9-GFP cells presenting EIINFEKL in the absence or presence of selected γc-cytokines. After treatment for 24 hours, cells were washed and then stained for flow cytometric analysis. Intracellular staining for IL-13 and IL-21 was performed using PE-labeled primary Abs according to the manufacturer's instructions. Granzyme B, BCL-6, and BLIMP-1 intracellular staining was performed after culture for 3 days under the specified conditions.
Localization of thymic PLZF+ cells by immunofluorescence
Thymi were harvested from 8-week-old WT C57BL/6 mice and embedded in OCT compound (Sakura Finetech) for cryosectioning. Sections (8 μm) were postfixed in 4% paraformaldehyde for 10 minutes, permeabilized with Triton X-100 0.2%, blocked, and then stained with PLZF-specific primary Ab and secondary anti–rabbit Alexa Fluor-488. Sections were observed with Zeiss LSM500 confocal microscope and images were acquired at room temperature at a 25× magnification with an LCI Plan-Neofluor 25×/0.8 differential interference contrast microscope as described previously.19
Statistical analyses
P values were calculated using the paired Student t test or ANOVA according to the experimental setup.
Results
Basic components of a culture system for analysis of CD8 T-cell positive selection and differentiation
We posited that a most parsimonious model should involve 3 components: DP thymocytes, peptide-coated stromal cells, and cytokines (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). As a source of DP thymocytes, we used OT-I–transgenic mice because peptides inducing positive or negative selection of OT-I thymocytes have been identified previously.21-24 We elected to use OP9 cells as stromal cells because this cell line is available in unlimited amounts and these cells express cell-surface levels of H2Kb similar to those of primary TECs (CD326+, also known as Epcam+, CD45−) harvested from C57BL/6 thymi (supplemental Figure 1B). After mild acid elution of endogenous MHC I–associated peptides, OP9 cells were pulsed with graded concentrations of synthetic peptides. In contrast to the influenza-derived H2Db-binding peptide (QDIENMETI), the 4 peptides recognized by the OT-I TCR bound to H2Kb on OP9 cells, as evidenced by the increased abundance of H2Kb at the cell surface of peptide-pulsed cells (supplemental Figure 1C). In the presence of the 4 H2Kb-binding peptides, H2Kb levels reached a plateau at 1 μg/mL (supplemental Figure 1C). Cocultures of DP thymocytes with unpulsed or peptide-pulsed OP9 cells were supplemented with γc-cytokines (rIL-2, rIL-4, rIL-7, rIL-9, rIL-15, rIL-21) or rIL-13 (closely related to rIL-4) for 3 days before analysis of DP and CD8 T cells.
Characterization of OT-I DP thymocyte subsets
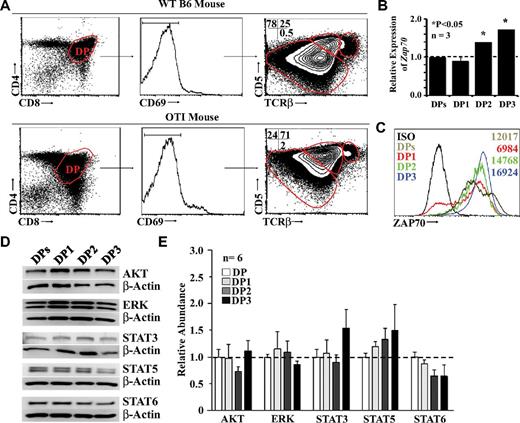
During thymocyte development, TCR signals up-regulate CD5, which in turn negatively regulates TCR signaling.25 By examining the surface expression intensities of TCR and CD5, Saini et al noted 3 distinct CD4+CD8+ DP populations in WT C57Bl/6 mice that they referred to as DP1 (TCRloCD5lo), DP2 (TCRintCD5hi), and DP3 (TCRhiCD5int).5,26 The same subsets of DP thymocytes were found in OT-I transgenic mice, but their distribution was different from that found in WT thymi. More specifically, the representation of DP subsets among WT nonselected DP thymocytes (CD4+CD8+CD69−) was as follows: DP1, 78%; DP2, 25%; and DP3, 0.5% (Figure 1A top panel). In the case of transgenic OT-I mice, the proportions of DP1, DP2, and DP3 thymocytes were 24%, 71%, and 2%, respectively (Figure 1A bottom panel). One additional hallmark differentiating the DP subsets is the expression level of the ζ-chain (TCR)–associated protein kinase of 70 kDa (ZAP70).5 We showed by qPCR (Figure 1B) and intracellular protein staining (Figure 1C) that the number of Zap70 transcripts and ZAP70 protein increased progressively from the DP1 to the DP3 developmental stages.
Characterization of fractionated OT-I DP thymocytes. (A) Representative flow cytometric analysis of WT (top panel) or OT-I (bottom panel) DP thymocytes. (B) Representative qPCR analysis of ZAP70 transcripts in total or fractionated OT-I DP thymocytes. Data depict transcript abundance relative to WT DP thymocytes. We tested 3 mice per group. *P < .05. (C) Representative flow cytometric analysis of ZAP70 intracellular staining performed on total or fractionated OT-I DP thymocytes. The isotype control is shown in black, total DPs in brown, DP1 in red, DP2 in green, and DP3 in blue. Numbers indicate the mean fluorescence intensity. Data shown are representative of 3 separate experiments. (D) Representative Western blot analysis of signaling molecules involved in positive selection. (E) Densitometry quantification of all Western blots performed. Six mice per group were tested.
Characterization of fractionated OT-I DP thymocytes. (A) Representative flow cytometric analysis of WT (top panel) or OT-I (bottom panel) DP thymocytes. (B) Representative qPCR analysis of ZAP70 transcripts in total or fractionated OT-I DP thymocytes. Data depict transcript abundance relative to WT DP thymocytes. We tested 3 mice per group. *P < .05. (C) Representative flow cytometric analysis of ZAP70 intracellular staining performed on total or fractionated OT-I DP thymocytes. The isotype control is shown in black, total DPs in brown, DP1 in red, DP2 in green, and DP3 in blue. Numbers indicate the mean fluorescence intensity. Data shown are representative of 3 separate experiments. (D) Representative Western blot analysis of signaling molecules involved in positive selection. (E) Densitometry quantification of all Western blots performed. Six mice per group were tested.
In addition to ZAP70, positive selection and CD8 T-cell differentiation depend on the following key molecules: ERKs, the serine-threonine kinase AKT, and STATs.6,12,27,28 We found no significant differences at the transcript or protein level in the expression of these signaling molecules in DP subsets (Figure 1D-E).
TCR-mediated modulation of cytokine receptors on DP thymocytes
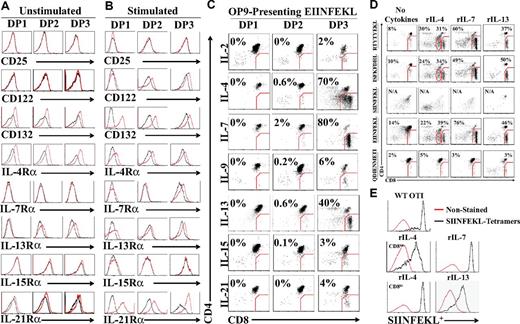
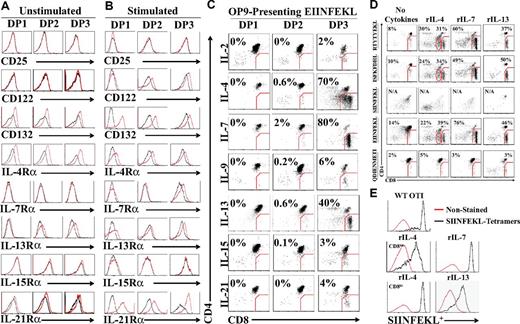
At the preselection stage, DP thymocytes are unresponsive to intrathymic IL-7 unless TCR signaling occurs and leads to de novo cell-surface expression of the IL-7 receptor-α (IL-7Rα) chain.6,29 Based on this observation, we investigated the expression profile of all γc-cytokine receptors (except for IL-9R because of the lack available Ab) on DP subsets using EIINFEKL-presenting OP9 cells. We also analyzed the expression of IL-13Rα (which is not a γc-cytokine) because, although IL-13 does not signal through the type I IL-4R (composed of IL-4Rα and IL-4Rγ), both IL-4 and IL-13 can signal through the type II IL-4R (IL-4Rα and IL-13Rα).30 Before TCR stimulation, fractionated DP1-3 thymocytes expressed γc (CD132) and the IL-4Rα chain (CD124). Minimal expression of the IL-13Rα chain was also detected on DP2 and DP3 thymocytes (Figure 2A). Presentation of EIINFEKL-H2Kb complexes to OT-I DP thymocytes up-regulated γc on all DP fractions and de novo expression of the IL-2/IL-15Rβ-chain (CD122) on both DP2 and DP3 cells. Conversely, the IL-7Rα chain was only detected on DP3 cells. Unexpectedly, we found that IL-21Rα was strongly up-regulated on the surface of all DP thymocyte fractions (Figure 2B). We next sought to evaluate the biologic relevance of these observations by testing the ability of each γc cytokine and IL-13 to enable the differentiation of OT-I DP thymocytes to the SP stage. Fractionated OT-I DP thymocytes cocultured for 3 days with EIINFEKL-presenting OP9 cells differentiated to CD8 SP T cells in the presence of rIL-4, rIL-7, and rIL-13 treatments, albeit with different efficiencies (Figure 2C). rIL-2, rIL-9, rIL-15, and rIL-21 did not support differentiation to the CD8 SP stage.
Cytokine receptor modulation and response during positive selection of OT-I DP thymocytes. (A-B) Representative flow cytometric analysis of cytokine receptor chains expression on fractionated DP1, DP2, and DP3 thymocytes in the presence of unpulsed (A) or EIINFEKL-pulsed (B) OP9 cells. Black lines represent the isotype control and the red line represents the test condition. (C) Generation of OT-I CD8 SP T cells in cocultures of DP thymocytes with EIINFEKL-pulsed OP9 cells in the presence of different cytokines (2 ng/mL for all cytokines except for rIL-21, which was used at 10 ng/mL). (D) Representative flow cytometric analysis of ex vivo developed OT-I CD8 SP T cells after culture with OP9 cells pulsed with various peptides in the presence of rIL-4, rIL-7, or rIL-13 (2 ng/mL). (E) SIINFEKL-H2Kb tetramer staining of OT-I CD8 SP T cells generated in the presence of different cytokines (2 ng/mL). Red line represents unstained cells and the black line represents tetramer-stained cells. All data shown are representative of 3 separate experiments.
Cytokine receptor modulation and response during positive selection of OT-I DP thymocytes. (A-B) Representative flow cytometric analysis of cytokine receptor chains expression on fractionated DP1, DP2, and DP3 thymocytes in the presence of unpulsed (A) or EIINFEKL-pulsed (B) OP9 cells. Black lines represent the isotype control and the red line represents the test condition. (C) Generation of OT-I CD8 SP T cells in cocultures of DP thymocytes with EIINFEKL-pulsed OP9 cells in the presence of different cytokines (2 ng/mL for all cytokines except for rIL-21, which was used at 10 ng/mL). (D) Representative flow cytometric analysis of ex vivo developed OT-I CD8 SP T cells after culture with OP9 cells pulsed with various peptides in the presence of rIL-4, rIL-7, or rIL-13 (2 ng/mL). (E) SIINFEKL-H2Kb tetramer staining of OT-I CD8 SP T cells generated in the presence of different cytokines (2 ng/mL). Red line represents unstained cells and the black line represents tetramer-stained cells. All data shown are representative of 3 separate experiments.
Previous fetal thymus organ culture–based studies have shown that the SIINFEKL peptide causes negative selection of OT-I thymocytes, whereas positive selection of OT-I can be induced by 3 peptides: EIINFEKL (a variant of the SIINFEKL peptide) and 2 natural peptides derived from the F-acting capping protein A (ISFKFDHL) and β-catenin (RTYTYEKL).21,22,24 Therefore, we tested the ability of these 4 peptides to support the differentiation of DP3 OT-I thymocytes into CD8 SP T cells in our culture systems. First, we performed a dose-response curve for each cytokine and found that the optimal concentration for rIL-4, rIL-7, and rIL-13 was 2 ng/mL (supplemental Figure 2). The H2-Db–binding peptide QDIENMETI was included as a negative control. In the absence of cytokines, a small percentage of CD8 T cells emerged from OT-I DP3 (Figure 2D), but not DP1 or DP2 (supplemental Figure 3A). Likewise, trace amounts of SP CD8 T cells could be obtained after culture of WT C57BL/6 DP3 in the absence of peptide-pulsed OP9 cells (supplemental Figure 3B). This basal peptide- or cytokine-independent production of CD8 SP T cells was absent when DP3 thymocytes were harvested from B2m−/− mice (supplemental Figure 3B), and was therefore likely because of in vitro completion of differentiation initiated in vivo by rare DP3 cells. Nonetheless, the salient finding is that supplementing the OP9/DP3 coculture with rIL-4, rIL-7, or rIL-13 triggered efficient CD8 T-cell development from DP3 thymocytes with all 3 positively selecting peptides (EIINFEKL, RTYTYEKL, and ISFKFDHL; Figure 3D). SP CD8 T cells generated in these conditions were confirmed to be SIINFEKL specific by tetramer-binding analysis (Figure 2E). As expected, the agonist peptide SIINFEKL negatively selected DP thymocytes, whereas the H2-Db influenza–derived peptide QDIENMETI was ineffective at supporting SP CD8 T-cell differentiation (Figure 2D). Interestingly, supplementing positively selected DP3 thymocytes with rIL-4 triggered the appearance of 2 distinct T-cell populations based on their CD8 coreceptor cell-surface expression intensity, which will be referred to hereafter as CD8int and CD8hi T cells (Figure 2C-E). It must be emphasized that pre-culture enrichment for DP3 thymocytes dramatically enhances the efficiency of SP CD8 T-cell production, which was nearly undetectable (0.6%-1.5%) when starting with unfractionated DP thymocytes (supplemental Figure 4). These results show that 3 cytokines (rIL-4, rIL-7, and rIL-13) can support the differentiation of DP3 thymocytes into SP CD8 T cells after positive selection by any of the 3 peptides tested. IL-4 has the unique ability to generate a population of SP CD8 T cells that is bimodal with regard to CD8 expression (CD8int and CD8hi).
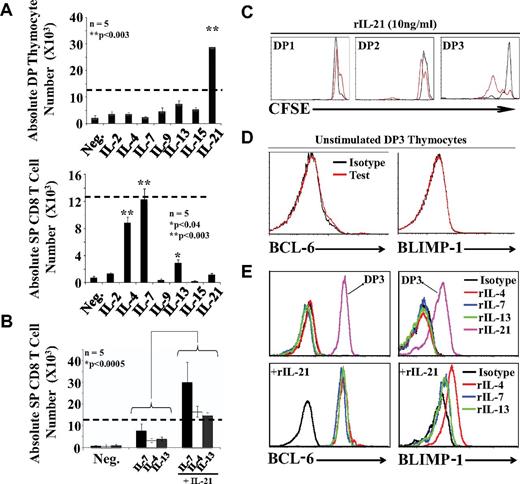
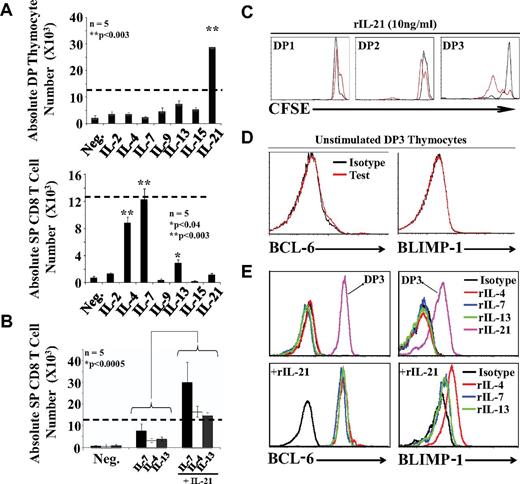
IL-21 has a unique ability to expand DP thymocytes during positive selection. (A) Absolute numbers of DP (top panel) and SP (bottom panel) CD8 thymocytes after culture of 12.5 × 103 DP3 OT-I thymocytes with EIINFEKL-pulsed OP9 cells supplemented with cytokines. Dashed lines represent the number of cells seeded per well at time 0. (B) Synergistic effect of IL-21 on CD8 T-cell differentiation. Five mice per group were tested. (C) CFSE content in DP1/DP2/DP3 thymocytes supplemented with 10 ng/mL of rIL-21 in the absence (black line) or presence (red line) of EIINFEKL-pulsed OP9 cells. (D) Representative flow cytometric analysis of intracellular BCL-6 and BLIMP-1 expression in unstimulated DP3 thymocytes. (E) Representative flow cytometric analysis of intracellular BCL-6 and BLIMP-1 expression in cocultures supplemented with various cytokines (2 ng/mL except for rIL-21, which was used at 10 ng/mL). Histograms are gated on SP CD8 T cells except for the 2 histograms gated on DP3 cells in the top panels. All data shown are representative of 3 separate experiments.
IL-21 has a unique ability to expand DP thymocytes during positive selection. (A) Absolute numbers of DP (top panel) and SP (bottom panel) CD8 thymocytes after culture of 12.5 × 103 DP3 OT-I thymocytes with EIINFEKL-pulsed OP9 cells supplemented with cytokines. Dashed lines represent the number of cells seeded per well at time 0. (B) Synergistic effect of IL-21 on CD8 T-cell differentiation. Five mice per group were tested. (C) CFSE content in DP1/DP2/DP3 thymocytes supplemented with 10 ng/mL of rIL-21 in the absence (black line) or presence (red line) of EIINFEKL-pulsed OP9 cells. (D) Representative flow cytometric analysis of intracellular BCL-6 and BLIMP-1 expression in unstimulated DP3 thymocytes. (E) Representative flow cytometric analysis of intracellular BCL-6 and BLIMP-1 expression in cocultures supplemented with various cytokines (2 ng/mL except for rIL-21, which was used at 10 ng/mL). Histograms are gated on SP CD8 T cells except for the 2 histograms gated on DP3 cells in the top panels. All data shown are representative of 3 separate experiments.
IL-21 is a potent mitogen for positively selected DP thymocytes
We were intrigued by the fact that although rIL-21 did not support the differentiation of SP CD8 thymocytes, the expression of IL-21Rα was strongly up-regulated on DP thymocytes undergoing positive selection (Figure 2B). We therefore sought to evaluate what might be the role of the de novo appearance of IL-21Rα during positive selection. To this end, we evaluated the effect of γc-cytokines and IL-13, not only on the generation of SP CD8 T cells, but also on the number of DP cells. DP3 OT-I thymocytes were cocultured with EIINFEKL-pulsed OP9 cells supplemented with individual γc cytokines. Again, SP CD8 T cells were found only in cultures supplemented with rIL-4, rIL-7, or rIL-13 (Figure 3A bottom panel). The significant finding here is that rIL-21 showed a unique ability to expand DP3 cells (Figure 3A top panel). On day 3, in the presence of rIL-21, the DP3 population had increased by approximately 3-fold. To determine whether rIL-21 can synergize with other cytokines for the production of SP CD8 T cells, we combined rIL-21 with differentiation-inducing cytokines (rIL-4, rIL-7, or rIL-13). rIL-21 synergized with the 3 cytokines because it increased by approximately 3-fold the number of SP CD8 T cells compared with cultures containing differentiation inducing cytokines but no rIL-21 (Figure 3B). The mitogenic effect of rIL-21 on DP3 thymocytes was further confirmed by analysis of CFSE-labeled cells (Figure 3C).
Strong evidence suggests IL-21 promotes the generation of long-lived memory CD8 T cells via STAT3-dependent up-regulation of BCL-6 and BLIMP-1.31 In the present study, we detected no expression of BCL-6 or BLIMP-1 in untreated DP3 thymocytes (Figure 3D) or in CD8 SP T cells generated in cocultures supplemented with rIL-4, rIL-7, or rIL-13 (Figure 3E top panels). However, we found a strong induction of both transcriptional repressors in DP3 thymocytes cocultured with EIINFEKL-pulsed OP9 cells and rIL-21 (Figure 3E top panels). Furthermore, admixing rIL-21 with differentiation-inducing cytokines led to a weak up-regulation of BLIMP-1 (in particular with rIL-4) and a major up-regulation of BCL-6 in SP CD8 T cells (Figure 3E bottom panels). We conclude that rIL-21 has a unique ability to induce production of BCL-6 and, to a lesser extent, BLIMP-1 in positively selected DP3 and SP CD8 T cells. We speculate that up-regulation of BCL-6 and BLIMP-1 is undetectable in unstimulated ex vivo harvested DP3 cells (Figure 3D) because it is dependent on continuous cell-cell contact or cytokine signaling and is therefore lost during cell preparation.32
IL-4 generates distinct populations of CD8 T cells
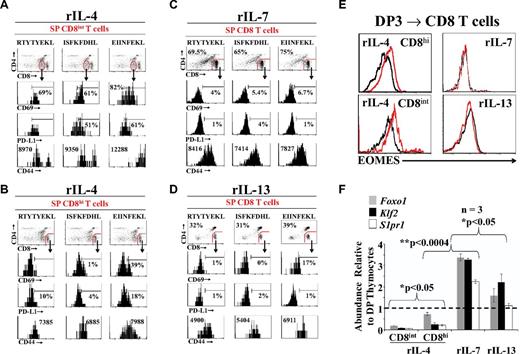
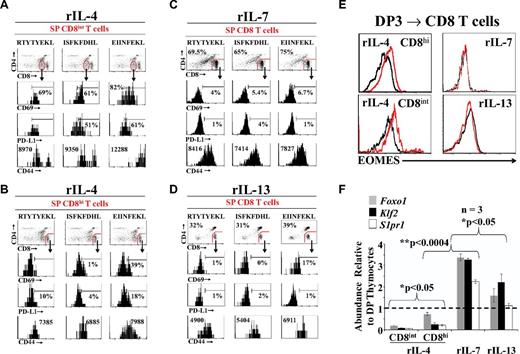
Prompted by the observation that rIL-4, but not rIL-7 or rIL-13, generated 2 distinct populations of SP CD8 T cells (Figure 2D-E), we wished to determine to what extent cytokines differentially influenced the differentiation of SP CD8 T cells. We examined the phenotype of OT-I SP CD8 T cells selected by OT-I–specific peptides in the presence of rIL-4, rIL-7, or rIL-13. Relative to CD8hi T cells generated in the presence of rIL-4, rIL-7, and rIL-13 (Figure 4C-D) and to freshly harvested OT-I CD8 SP thymocytes (supplemental Figure 6B), CD8int T cells induced by rIL-4 expressed higher levels of CD69, PD-L1, and CD44 (Figure 4A-B). The CD8intCD69hiPD-L1hiCD44hi phenotype is typical of innate polyclonal TCRαβ CD8 T cells15,16 and was more conspicuous in the presence of EIINFEKL than other peptides. These CD8intCD69hiPD-L1hiCD44hi cells did not bind TL tetramers (supplemental Figure 6A), suggesting that they do not express CD8αα homodimers. Interestingly, rIL-4 was dominant over rIL-7: when they differentiated in the presence of both rIL-4 and rIL-7, SP CD8 T cells had a phenotype similar to those cultured in the presence of IL-4 alone (supplemental Figure 5). Furthermore, all in vitro generated CD8 T cells expressed Runx3, a hallmark of the CD8 lineage (supplemental Figure 7A) and were functional, as evidenced by their production of granzyme B after CD3/CD28 stimulation in vitro (supplemental Figure 7B).
Characterization ofex vivogenerated OT-I CD8 SP T cells. (A-B) Representative flow cytometric analysis of OT-I CD8int (A) or CD8hi (B) SP T cells phenotype on rIL-4 treatment. (C-D) Representative phenotypic analysis of OT-I CD8 SP T cells generated in the presence of rIL-7 (C) or rIL-13 (D). (E) Representative flow cytometric analysis of EOMES intracellular staining of OT-I CD8 SP T cells generated from the positive selection of DP3 thymocytes. The black line represents the isotype control and the red line represents EOMES staining. (F) qPCR analysis of Foxo1, Klf2, and S1pr1expression on ex vivo generated OT-I CD8 SP T cells. Data depict transcript abundance relative to untreated DP thymocytes. Three mice per group were tested. *P < .05; **P < .0004. For panels A through F, data shown are representative of 3 separate experiments.
Characterization ofex vivogenerated OT-I CD8 SP T cells. (A-B) Representative flow cytometric analysis of OT-I CD8int (A) or CD8hi (B) SP T cells phenotype on rIL-4 treatment. (C-D) Representative phenotypic analysis of OT-I CD8 SP T cells generated in the presence of rIL-7 (C) or rIL-13 (D). (E) Representative flow cytometric analysis of EOMES intracellular staining of OT-I CD8 SP T cells generated from the positive selection of DP3 thymocytes. The black line represents the isotype control and the red line represents EOMES staining. (F) qPCR analysis of Foxo1, Klf2, and S1pr1expression on ex vivo generated OT-I CD8 SP T cells. Data depict transcript abundance relative to untreated DP thymocytes. Three mice per group were tested. *P < .05; **P < .0004. For panels A through F, data shown are representative of 3 separate experiments.
The generation of innate polyclonal CD8 T cells is dependent on up-regulation of eomesodermin (EOMES) by IL-4. Therefore, when mutant mice possessing high numbers of innate CD8 T cells (Klf2−/−, Itk−/−, or Id3−/−) were crossed with IL4r-deficient mice, they did not up-regulate Eomes nor develop innate CD8 T cells.15,33 We found that the hallmark of innate CD8 T cells, up-regulation of EOMES, was induced in SP CD8 T cells positively selected in the presence of rIL-4, but not of rIL-7 or rIL-13 (Figure 4E).
TCR signaling by pMHC complexes induces the phosphorylation of AKT, leading to forkhead box O1 (FOXO1) sequestration in the cytoplasm and transient expression of CD69.7 This in turns directly represses the transcriptional activity of Kruppel-like factor 2 (Klf2) and GPCR sphingosine-1-phosphate receptor 1 (S1pr1), resulting in thymic retention of developing thymocytes.34-36 Termination of TCR engagement and differentiation to the SP stage halt AKT activation, thereby inducing FOXO1-dependent transcription of Klf2 and S1pr1.7 The end result is the down-regulation of cell-surface CD69, which primes SP T cells for thymic export. In the present study, we observed that Foxo1, Klf2, and S1pr1 transcripts were up-regulated in SP CD8 T cells that differentiated in the presence of rIL-7 and, to a lesser extent, rIL-13 (Figure 4F). In contrast, these 3 transcripts were repressed by rIL-4, particularly in the SP CD8int subset (Figure 4F). We conclude that whereas rIL-7 and rIL-13 generate classic SP CD8 T cells, rIL-4 generates 2 distinct populations of CD8 T cells. One subset of rIL-4–induced T cells is CD8intPD-L1hiCD44hi (innate CD8 T cells), and the other is CD8hiPD-L1−CD44int (like classic CD8 T cells). In contrast to classic (ie, rIL-7–induced) T cells, the 2 rIL-4–dependent populations are EOMES+CD69+Foxo1loKlf2loS1pr1lo. The CD69+Foxo1loKlf2loS1pr1lo phenotype of IL-4–induced CD8 T cells suggests that they are ill equipped for thymus emigration compared with classic rIL-7–induced CD8 T cells.
TCR- versus cytokine-dependent signaling events
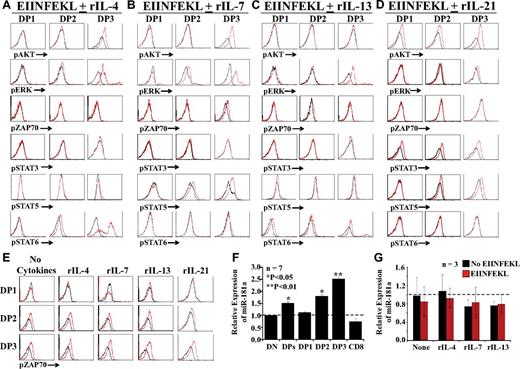
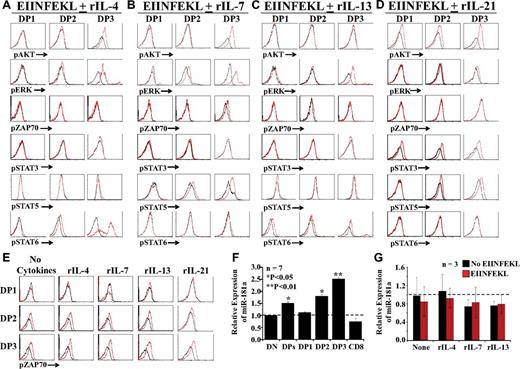
Up-regulation of miR-181a and phosphorylation of AKT, ERK, ZAP70, and STAT are instrumental in positive selection and differentiation of CD8 T cells.37 We first analyzed the phosphorylation of signaling molecules in OT-I DP thymocytes cultured with peptide-pulsed OP9 cells in medium supplemented with rIL-4, rIL-7, rIL-13, or rIL-21. Treatment with IL-4 induced a slight activation of STAT6 in DP2 thymocytes and a strong activation of AKT, ERK, and STAT6 in DP3 thymocytes (Figure 5A). In the presence of IL-7, levels of pERK and pSTAT5 in DP2 were slightly increased in DP2 thymocytes, whereas pAKT, pERK, and pSTAT5 were conspicuously more abundant in DP3 thymocytes (Figure 5B). Treatment with rIL-13 induced a weak phosphorylation of ERK and a slight activation of STAT6 in DP3 thymocytes, but had no effect on other DP subsets (Figure 5C). Conversely, IL-21 induced STAT3 phosphorylation in all DP fractions and phosphorylation of AKT and STAT5 in DP3 thymocytes (Figure 5D). Consistent with the absence of IL-2Rα and IL-15Rα and despite the presence of IL-2/15Rβ on DP3 thymocytes (Figure 2A-B), even high concentrations of IL-2 or IL-15 did not promote DP3 differentiation to the CD8 SP stage (supplemental Figure 8A). The inability of IL-9 to induce pSTAT3 and pSTAT5 in DP thymocytes (supplemental Figure 8B) shows that DP3 thymocytes are unresponsive to IL-9 (Figure 3A), presumably because they do not express Il-9R.
TCR- versus cytokine-dependent signaling events. (A-D) Representative flow cytometric analysis of phosphoproteins in fractionated DP thymocytes cultured for with peptide-pulsed OP9 cells for 24 hours in medium supplemented with rIL-4, rIL-7, rIL-13, or rIL-21 (red histograms) or no cytokines (black histograms). (E) DP thymocytes were cultured in medium containing rIL-4, rIL-7, rIL-13, rIL-21, or no cytokines with (red) or without (black) EIINFEKL-pulsed OP9 cells. For panels A through E, 3-6 mice per group were tested in separate experiments. (F) qPCR analysis of miR-181a expression in freshly harvested thymocyte subsets from OT-I mice. Data depict transcript abundance relative to DN thymocytes. (G) qPCR analysis of miR-181a in DP3 thymocytes cultured for 24 hours in cytokine-supplemented medium in the presence or absence of peptide-pulsed OP9 cells. In panels F and G, 3-7 mice per group were tested. *P < .05; **P < .01.
TCR- versus cytokine-dependent signaling events. (A-D) Representative flow cytometric analysis of phosphoproteins in fractionated DP thymocytes cultured for with peptide-pulsed OP9 cells for 24 hours in medium supplemented with rIL-4, rIL-7, rIL-13, or rIL-21 (red histograms) or no cytokines (black histograms). (E) DP thymocytes were cultured in medium containing rIL-4, rIL-7, rIL-13, rIL-21, or no cytokines with (red) or without (black) EIINFEKL-pulsed OP9 cells. For panels A through E, 3-6 mice per group were tested in separate experiments. (F) qPCR analysis of miR-181a expression in freshly harvested thymocyte subsets from OT-I mice. Data depict transcript abundance relative to DN thymocytes. (G) qPCR analysis of miR-181a in DP3 thymocytes cultured for 24 hours in cytokine-supplemented medium in the presence or absence of peptide-pulsed OP9 cells. In panels F and G, 3-7 mice per group were tested. *P < .05; **P < .01.
Under our culture conditions, the presence or absence of cytokines did not change the levels of pZAP70 in DP thymocytes (Figure 5A-D). We therefore performed a separate series of experiments in which 2 variables were manipulated: the presence or absence of cytokines and the presence or absence of peptide-pulsed OP9 cells. We found that the amount of pZAP70 was increased in the presence of peptide-pulsed OP9 cells but unaffected by the presence or absence of cytokines (Figure 5E). The importance of miR-181a in positive selection stems from its ability to repress several phosphatases downstream of TCR signaling and thereby to increase the sensitivity to pMHC complexes.38 qPCR analysis on thymocyte subsets showed that miR-181a up-regulation was initiated at the DP2 stage, reached a maximum in DP3 thymocytes, and was down-regulated in SP CD8 T cells (Figure 5F). Expression of miR-181a remained unchanged when DP3 thymocytes were cultured for 24 hours with unpulsed or peptide-pulsed OP9 cells with or without cytokines (Figure 5G). We conclude that in DP3 thymocytes, activation of ZAP70 and expression of miR-181a are unaffected by cytokines. In addition, ZAP70 phosphorylation, but not miR-181a expression, remains dependent on TCR signals.
Expression of chemokine receptors
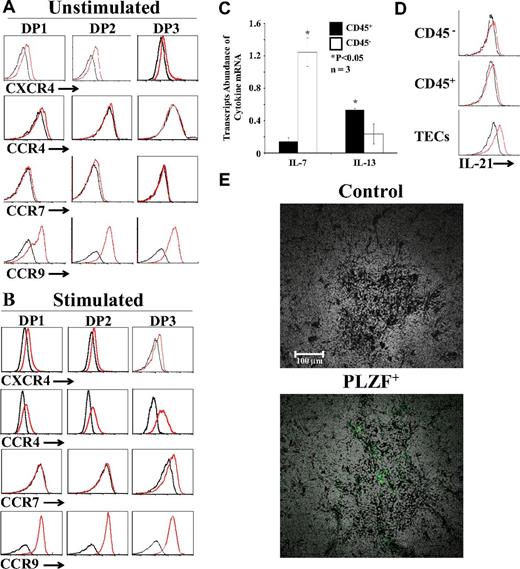
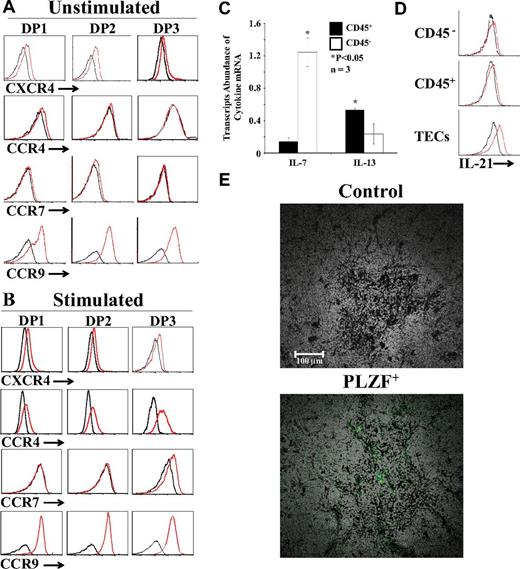
Directional migration of thymocytes during T-cell development is regulated by chemokines and chemokine receptors. Positive selection of DP thymocytes is correlated wi,th the down-regulation of CXCR4 expression intensity and the up-regulation of CCR4 and CCR7 with enhanced migration toward CCR7 and CCR9 ligands.6,7,39 Modulation of chemokine receptors after positive selection promotes the migration of thymocytes to the medulla, where they can complete their differentiation and emigrate toward the periphery. To evaluate whether TCR signals were sufficient to modulate chemokine receptor expression, we analyzed cell-surface expression of CXCR4, CCR4, CCR7, and CCR9 on fractionated DPs cultured with unpulsed or peptide-pulsed OP9 cells (without cytokine supplementation). Unstimulated DP thymocytes were CXCR4loCCR4−CCR7−CCR9+ (Figure 6A). TCR stimulation with the EIINFEKL peptide in the absence of γc cytokines did not modulate CXCR4 expression but did induce de novo expression of CCR4 on all DP fractions and of CCR7 on DP3 thymocytes (Figure 6B). In addition, TCR stimulation induced a weak yet significant up-regulation of CCR9 (Figure 6B). We found that the addition of IL-4, rIL-7, or IL-13 during TCR stimulation did not modulate chemokine receptor expression (supplemental Figure 9). Based on these data, we conclude that positive selection, but not cytokines, regulates the expression of CCR4, CCR7, and CCR9.
Expression of chemokine receptors and identification of cytokine-producing cells. (A-B) Representative flow cytometric analysis of CXCR4, CCR4, CCR7, and CCR9 on fractionated OT-I DPs cultured with unpulsed (A) or EIINFEKL-pulsed (B) OP9 cells without cytokine supplementation. Black lines represent isotype controls. (C) qPCR analysis of IL-7 and IL-13 transcripts in CD45+ (black bars) and CD45− (open bars) cells sorted from the thymi of C57BL/6 mice; transcript abundance in CD45+ versus CD45− cells was different. *P < .05. (D) Flow cytometric analysis of IL-21 expression in hematopoietic (CD45+), nonhematopoietic (CD45−), and epithelial (EpCAM+CD45−) thymic cells. (E) Immunolocalization of PLZF+ cells in the thymus. Thymi from 8-week-old WT C57BL/6 mice were fixed in 10% formalin, permeabilized with Triton X-100 0.2%, and mounted in paraffin. Sections were stained with PLZF-specific primary Ab and secondary anti–rabbit Alexa Fluor 488, and then scanned using the NanoZoomer Digital Pathology system and NPD.scan Version 2.3.4 software (Hamamatsu) as described previously (40×/1.3 numerical aperture oil objective).19 Data are representative of 3 separate experiments.
Expression of chemokine receptors and identification of cytokine-producing cells. (A-B) Representative flow cytometric analysis of CXCR4, CCR4, CCR7, and CCR9 on fractionated OT-I DPs cultured with unpulsed (A) or EIINFEKL-pulsed (B) OP9 cells without cytokine supplementation. Black lines represent isotype controls. (C) qPCR analysis of IL-7 and IL-13 transcripts in CD45+ (black bars) and CD45− (open bars) cells sorted from the thymi of C57BL/6 mice; transcript abundance in CD45+ versus CD45− cells was different. *P < .05. (D) Flow cytometric analysis of IL-21 expression in hematopoietic (CD45+), nonhematopoietic (CD45−), and epithelial (EpCAM+CD45−) thymic cells. (E) Immunolocalization of PLZF+ cells in the thymus. Thymi from 8-week-old WT C57BL/6 mice were fixed in 10% formalin, permeabilized with Triton X-100 0.2%, and mounted in paraffin. Sections were stained with PLZF-specific primary Ab and secondary anti–rabbit Alexa Fluor 488, and then scanned using the NanoZoomer Digital Pathology system and NPD.scan Version 2.3.4 software (Hamamatsu) as described previously (40×/1.3 numerical aperture oil objective).19 Data are representative of 3 separate experiments.
Features of cytokine-producing cells
The results of the present study show that 4 cytokines influenced the quantity and quality of SP CD8 thymocytes in our system: rIL-4, rIL-7, rIL-13, and rIL-21. In the thymus, IL-7 is produced by TECs, particularly those located at the corticomedullary junction and in the medulla, whereas IL-4 is produced exclusively by PLZF+ NKT cells.15,40,41 We were unable to detect IL-4 by immunohistochemistry or IL-4 transcripts by in situ hybridization on thymus sections (data not shown). However, we were able to localize PLZF+ cells by immunohistochemistry on the thymi of WT C57BL/6. We found that PLZF+ cells (the source of IL-4) are rare and located in the medulla (Figure 6E). We were unable to detect IL-13 protein by flow cytometry, but, as expected,42 IL-13 transcripts were present at least in CD45+ thymic cells (Figure 6C). Flow cytometric analysis on primary thymic cell populations revealed the presence of IL-21 in TECs (Figure 6D). The notion that IL-21 is produced by TECs supports the idea that the effect of IL-21 on positively selected DP thymocytes is physiologically relevant. The fact that IL-4 is produced only by rare PLZF+ cells located in the medulla must be contrasted with the wider distribution (cortex and medulla) and greater abundance (mainly in the medulla) of IL-7–producing TECs.40,41 The differential distribution and abundance of cells producing IL-4 versus IL-7 must be taken into account when discussing the role of these cytokines in CD8 T-cell differentiation (see “Discussion”).
Injection of rIL-21 increases the number of DP thymocytes in vivo
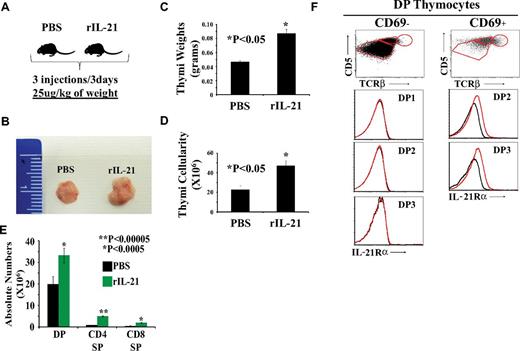
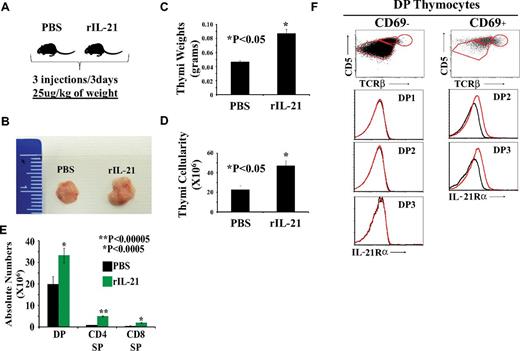
To assess the effect of IL-21 on thymopoiesis directly in vivo, we injected rIL-21 or PBS to old (13-14 months of age) WT C57Bl/6 mice. Littermates received 3 injections of PBS or rIL-21 at a dose of 25 μg/kg of body weight (Figure 7A). Thymi from mice in the rIL-21 group showed a noticeable increase in size (Figure 7B), weight (Figure 7C), and numbers of DP and SP thymocytes (Figure 7D-E). These data are consistent with the mitogenic effect of IL-21 on DP thymocytes in vitro (Figure 3) and demonstrate that a short course of treatment with rIL-21 is sufficient to increase the number of DP and SP thymocytes in vivo.
The effect of rIL-21 injection on thymi of old WT C57Bl/6 mice. (A) Schematic diagram of the experimental design. Three mice per group were tested. (B) Representative photograph of thymi derived from PBS- versus rIL-21–treated mice. (C-D) Thymic weight and cellularity. (E) Absolute numbers of DP, SP CD4, and CD8 T cells. (F) Analysis of IL-21Rα chain on the surface of preselected (CD69−) or postselected (CD69+) DP subsets. Data are representative of 3 separate experiments.
The effect of rIL-21 injection on thymi of old WT C57Bl/6 mice. (A) Schematic diagram of the experimental design. Three mice per group were tested. (B) Representative photograph of thymi derived from PBS- versus rIL-21–treated mice. (C-D) Thymic weight and cellularity. (E) Absolute numbers of DP, SP CD4, and CD8 T cells. (F) Analysis of IL-21Rα chain on the surface of preselected (CD69−) or postselected (CD69+) DP subsets. Data are representative of 3 separate experiments.
We also assessed the expression of IL-21R on preselected (CD69−) versus postselected (CD69+) DP thymocytes derived from WT C57Bl/6 mice (Figure 7F). None of the CD69− DP1, DP2, or DP3 subsets expressed IL-21R. As expected, immature DP1 thymocytes contained no CD69+ elements. In contrast, the DP2 CD69+ (capable of CD4 differentiation) and DP3 CD69+ (capable of CD8 differentiation) subsets expressed IL-21R on their cell surface (Figure 7F). These results agree with our in vitro observations (Figure 2A-B): IL-21R is up-regulated on DP thymocytes only when they receive TCR signals.
Discussion
Using a novel in vitro system for studying positive selection and T-cell differentiation, we provide evidence herein that 4 cytokines have differential effects on the maturation of positively selected CD8 thymocytes: rIL-4, rIL-7, rIL-13, and rIL-21 (supplemental Figure 10). Our in vitro system generates SP CD8 T cells in only 3 days and is both simple and flexible because it requires only 3 readily accessible components: sorted CD69− DP3 thymocytes, peptide-pulsed OP9 cells, and a γc cytokine. A significant advantage of our system is that it enables high-throughput analyses. The data presented herein are based on approximately 1200 individual cultures and this number could be easily increased by 10-fold to study, for example, positive selection by individual peptides (or peptide mixtures) from a library.
As proposed by Park et al,6 in the present study, cytokines did not affect positive selection per se: up-regulation of miR-181a and pZAP70 were cytokine independent. As expected, rIL-7 promoted the differentiation of positively selected CD8 T cells by inducing phosphorylation of STAT5, AKT, and ERK. That rIL-13 can support differentiation of SP CD8 T cells was somewhat unexpected because IL-13 does not belong to the γc-cytokine family. However, we consider that the biologic importance of IL-13 in vivo remains doubtful because, compared with IL-7, IL-13 was less effective (probably because it did not activate AKT) and did not exhibit any nonredundant effect.
Like IL-7, IL-4 activated AKT and ERK but signaled through STAT6 rather than STAT5. That rIL-4 can promote the differentiation of CD8 thymocytes is consistent with its ability to induce the expression of RUNX3, the transcription factor that specifies CD8 lineage choice.6 The salient finding with IL-4 was its unique ability to support the generation of CD8intPD-L1hiCD44hiEOMES+ innate CD8 T cells. This observation is consistent with previous work showing that innate TCRαβ CD8 T cells are IL-4 dependent15 and that they diverge from classic CD8 T cells during or shortly after positive selection.16 The CD69+Foxo1loKlf2loS1pr1lo phenotype of IL-4–induced CD8 T cells suggests that their migration toward the medulla and their export from the thymus might be delayed relative to classic CD8 T cells. Considering that only 10% of TCRαβ CD8 thymocytes are innate T cells in vivo,16 the observation that rIL-4 dominated rIL-7 was surprising: in the presence of equimolar concentrations of rIL-4 and rIL-7, DP3 thymocytes generated the same proportion of innate CD8 T cells as in cultures containing only rIL-4. These observations can be reconciled considering the distribution of IL-4- and IL-7–producing cells in the thymus. IL-7 is produced by a rich network of TECs, particularly at the corticomedullary junction and in the medulla, whereas IL-4 is produced by rare PLZF+ NKT cells in the medulla. We therefore surmise that most, if not all, thymocytes are exposed to IL-7 but only a minority encounter IL-4–producing cells.
Because young IL-21Rα–deficient mice display normal proportions of thymocyte subsets,43 IL-21 has been assumed to have no effect on thymopoiesis. Therefore, the unique ability of IL-21 to expand positively selected DP3 is perhaps the most interesting observation emerging from this work. We detected IL-21 in TECs and found that IL-21Rα was strongly up-regulated during positive selection. IL-21 did not promote CD8 T-cell differentiation, probably because of its inability to activate ERK (the phosphorylation of which was induced by rIL-4, rIL-7, and rIL-13). However, IL-21 was the sole cytokine that was mitogenic for DP thymocytes; the number of DPs harvested after culture in the presence of rIL-21 was superior to the number of DP3s plated per well. The effect of IL-21 on DPs was correlated with its ability to activate STAT3 and thereby modulate the expression of 2 STAT3 targets, BCL-6 and BLIMP-1. rIL-21 up-regulated BLIMP-1 mildly and BCL-6 strongly. BCL-6 and BLIMP-1 are antagonistic transcription factors that function as a self-reinforcing genetic switch for cell-fate decisions in several lymphoid cell populations.31,44 In general, BLIMP-1 promotes terminal differentiation and BCL-6 sustains the survival and proliferation potential. Consistent with this, we noted in the present study that when used alone or together with differentiation-inducing cytokines, rIL-21 up-regulated BCL-6 to a greater extent than BLIMP-1. Further work is needed to discover the targets of BCL-6 in DP3 thymocytes undergoing positive selection. The magnitude of thymic output is much more closely correlated with the number of DPs than with the number of early thymic progenitors.45,46 Furthermore, we found that admixing rIL-21 with rIL-7 dramatically increased the number of SP CD8 T cells generated in culture and that a short course of treatment with rIL-21 was sufficient to increase the numbers of DP and SP thymocytes in vivo. Although its effect on human thymic output has never been investigated, rIL-21 has been used for other purposes and is well tolerated in humans at doses up to 100 μg/kg.47 The results of the present study therefore suggest that injection of rIL-21 alone or with rIL-7 might represent a promising and unforeseen strategy to enhance thymic function in subjects with age- or disease-related thymic atrophy.48-50
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Danièle Gagné for flow cytometry and cell sorting, Raphaëlle Lambert for the qPCR experiments, and the staff of the Institute for Research in Immunology and Cancer animal care facility for their assistance.
This work was supported by grant MOP 42384 from the Canadian Institutes of Health Research. M.R. holds a Canadian Institutes of Health Research fellowship and C.P. holds a Canada Research Chair in Immunobiology. The Institute for Research in Immunology and Cancer is supported in part by the Canadian Center of Excellence in Commercialization and Research, the Canada Foundation for Innovation, and the Fonds de la Recherche en Santé du Québec.
Authorship
Contribution: M.R. designed the study, carried out the experiments, analyzed the data, prepared the figures, and wrote the first draft of the manuscript; A.R., S.B., and J.R.V. performed the experiments and participated in the data analysis; C.P. designed the study, discussed the results, and wrote the manuscript; and all authors edited and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Claude Perreault, Institute for Research in Immunology and Cancer, Université de Montréal, PO Box 6128, Station Centre-ville, Montréal, QC, Canada H3C 3J7; e-mail: claude.perreault@umontreal.ca.