Key Points
CtBP1 and FA proteins interact together and are implicated in the regulation of the Wnt antagonist Dickkopf-1
Abstract
Fanconi anemia (FA) is a genetic disorder characterized by congenital abnormalities, bone marrow failure, and increased susceptibility to cancer. Of the fifteen FA proteins, Fanconi anemia group C (FANCC) is one of eight FA core complex components of the FA pathway. Unlike other FA core complex proteins, FANCC is mainly localized in the cytoplasm, where it is thought to function in apoptosis, redox regulation, cytokine signaling, and other processes. Previously, we showed that regulation of FANCC involved proteolytic processing during apoptosis. To elucidate the biological significance of this proteolytic modification, we searched for molecular interacting partners of proteolytic FANCC fragments. Among the candidates obtained, the transcriptional corepressor protein C-terminal binding protein-1 (CtBP1) interacted directly with FANCC and other FA core complex proteins. Although not required for stability of the FA core complex or ubiquitin ligase activity, CtBP1 is essential for proliferation, cell survival, and maintenance of chromosomal integrity. Expression profiling of CtBP1-depleted and FA-depleted cells revealed that several genes were commonly up- and down-regulated, including the Wnt antagonist Dickkopf-1 (DKK1). These findings suggest that FA and Wnt signaling via CtBP1 could share common effectors.
Introduction
Fanconi anemia (FA) is an autosomal and X-linked genetic syndrome characterized by progressive bone marrow failure, congenital malformations, and predispositions for leukemia and other forms of cancer.1,2 To date, 15 gene products have been linked to FA or FA-like diseases that, when mutated, cause several cellular phenotypes, including hypersensitivity to DNA interstrand crosslinks (reviewed in de Winter and Joenji2 and Deans and West3 ). Consequently, these proteins are thought to function in a canonical pathway known as the “FA pathway,” which is involved in genome surveillance, but the exact role of this pathway (or individual FA proteins) that could explain clinical features of the disease remains unknown.
Of all FA pathway components, the Fanconi anemia group C (FANCC) protein is the only protein principally located within the cytoplasm.4,5 Although some FANCC localizes to the nucleus, its role in the cytoplasm is still unclear. FANCC binds to several cytosolic enzymes involved in the metabolism of oxygen radicals,6,7 protein chaperones,8,9 cell cycle regulators,10 and signal transduction components.11 Mutational studies have indicated that FANCC performs multiple functions by acting independently in at least two signaling response pathways: DNA crosslink damage–induced pathways and cytokine-induced pathways.12,13 These results suggest that FANCC acts as a transducer of signals or as a relay protein to regulate cell stress responses. In fact, FANCC overexpression prevents or delays apoptosis in response to various stimuli, including cytokines and DNA-damaging agents.14,15 In addition, we previously showed that FANCC undergoes caspase-mediated proteolytic modification that results in cleaved protein fragments, which may inhibit its function as a suppressor of apoptosis or may activate some proapoptotic function.16 To identify the biological functions of these FANCC protein fragments, we performed protein interaction screens to identify novel binding partners of FANCC. C-terminal binding protein-1 (CtBP1), a repressor of Wnt signaling, was identified among the proteins that interacted with FANCC.
The CtBP family of proteins consists of two homologs, CtBP1 and CtBP2, which are involved in transcriptional regulation, cell cycle progression, cell survival, and regulation of mitotic fidelity.17-20 Both are widely expressed throughout development and are often expressed in overlapping patterns, which leads to functional redundancy.21 CtBPs have at least two distinct roles: in the cytoplasm, they function in the Golgi tubule-fissioning machinery; in the nucleus, they cooperate with DNA-binding transcription factors, such as Wnt/β-catenin/T-cell factor (TCF), in recruiting chromatin-modifying enzymes to promoters to achieve transcriptional corepression.17,22,23
Here, we report that CtBP1 interacts directly with FANCC and other FA pathway components, which are FANCA, FANCF, FANCG, and FANCL. We show that, similar to FA proteins, CtBP1 is essential for cell proliferation, cell survival, and chromosomal integrity. Analysis of DNA damage–induced monoubiquitination of FANCD2 revealed that CtBP1 is dispensable for activation of the FA pathway in this context. However, cells depleted of CtBP1 or FA proteins exhibit up- and down-regulation of common genes, including Dickkopf-1 (DKK1), a negative regulator of the Wnt pathway.
Materials and Methods
Yeast 2-hybrid screens and analyses
Yeast 2-hybrid screens and analyses using the Matchmaker 2-Hybrid System 3 (Clontech, Mountain View, CA) were performed according to the manufacturer’s instructions. FANCC complementary DNA (cDNA) sequences corresponding to the cleaved FANCC N-terminal and C-terminal fragments16 (FANCC1-306 and FANCC307-558) were amplified via polymerase chain reaction (PCR) and cloned into pGBKT7 as fusions with the Gal4 DNA-binding domain. FANCC constructs were transformed into AH109 yeast and were mated to the Y187 strain, which was pretransformed with cDNA libraries obtained from either HeLa cells or human fetal brain (Clontech). Yeast diploids were plated in dropout medium lacking tryptophan, leucine, histidine, and adenine. Three different screens were performed using either pGBKT7-FANCC1-306 or pGBKT7-FANCC307-558 constructs as bait. For yeast 2-hybrid analyses with FA proteins, all plasmids and methods were used as previously described.24
Cell lines and culture conditions
HEK293T, HeLa, PD20 (FA-D2), and PD220 (FA-A), and their complemented counterpart (gift from the Fanconi Anemia Research Fund) fibroblast cell lines were grown in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum and were incubated in 5% CO2 at 37°C.
DNA constructs and antibodies
All FA gene constructs used were described previously.24 The pRc/CMV-T7-CtBP1 plasmid was a gift from Dr. G. Chinnadurai (St. Louis University Health Sciences Center). The following antibodies were used: anti-FANCA (C-20, Santa Cruz Biotechnologies, Santa Cruz, CA, or Novus Biologicals, Littleton, CO); anti-FANCC (previously described,16 Novus Biologicals; and 8F3, a gift from Dr. M. Hoatlin, Oregon Health & Science University and Novus Biologicals); anti-FANCD2 and anti-FANCE (Novus Biologicals); anti-CtBP1 (Millipore, Billerica, MA, or BD Biosciences, San Jose, CA); anti-CtBP2 (Synaptic Systems, Göttingen, Germany); anti-RAD51 (H92, Santa Cruz Biotechnologies); anti-HA (12CA5, Roche Diagnostics, Indianapolis, IN); anti-cMyc (9E10, Santa Cruz Biotechnologies); anti-FLAG (M2, Sigma, St. Louis, MO); anti-green fluorescent protein (GFP) (B-2, Santa Cruz Biotechnologies); anti-T7 (Novagen, Madison, WI); anti-GAPDH (1D4, Novus Biologicals); anti-goat (Calbiochem, San Diego, CA), anti-mouse and anti-rabbit (Santa Cruz Biotechnologies); and donkey anti-rabbit Alexafluor 488 and anti-mouse Alexafluor 555 (Invitrogen, Burlington, ON). All animal procedures were performed according to protocols approved by the Animal Care Committee of Laval University, Québec City, QB, Canada.
Immunoprecipitations
Whole cell extracts were subjected to either immunoblot analysis or immunoprecipitation as described previously.24 Equal amounts of protein were incubated overnight at 4°C with 2 µg of antibodies and then incubated with Protein A/G agarose beads (Calbiochem), or protein G magnetic beads (Invitrogen). Immunoprecipitates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subjected to western blotting with specific antibodies, which are indicated in each figure.
shRNA and production of lentivirus
A four-plasmid (pRSV-Rev, pMDLg/pRRE, pMD2.G, and pLKO.1) expression system was used for lentiviral production. Different pLKO.1 plasmids carrying short hairpin RNAs (shRNAs) targeting FANCA (TRCN0000118982), FANCD2 (TRCN0000082840), CtBP1 (TRCN0000013738), or CtBP2 (TRCN0000013744) were purchased from Open Biosystems (Lafayette, CO). A lentiviral control vector, pLKO.1-scrambled was purchased from Addgene (Cambridge, MA). Lentiviral particles were produced via calcium phosphate–mediated transient transfection of the four plasmids into HEK293T cells. In each experiment, HeLa cells were transduced for 6 hours with filtered supernatant containing recombinant lentiviral particles. After transduction, the cells were cultured for 48 to 96 hours and analyzed for knockdown efficiencies (supplemental Figure 1). Proliferation, cell survival, and cell cycle assays in conjunction with western blot analyses revealed a critical point at 72 hours (3 days) post-transduction when cells were depleted of the target gene but were still viable. Thus, unless otherwise indicated, all experiments involving the use of knockdown cells were conducted at 72 hours post-transduction.
Cell growth, survival, and reporter assays
HeLa cells were exposed to appropriate lentiviral particles. Cell viability was determined each day after transduction via staining with an acetomethoxy derivate of calcein (calcein-AM), which was used in accordance with the manufacturer’s instructions (Molecular Probes, Burlington, ON, Canada). To obtain an accurate cell count, CountBright absolute counting beads (Invitrogen) were added to each sample, and data acquisitions were done according to the manufacturer’s instructions. For the luciferase reporter assays, transduced cells were transfected with the M50 Super 8× TOPflash TCF/lymphoid-enhancing factor (TCF/LEF) reporter plasmid (Addgene plasmid #12456; Addgene) or the M51 Super 8× FOPflash TCF/LEF mutant (Addgene plasmid #12457; Addgene) along with the renillia luciferase control plasmid (Promega, Madison, WI). Luciferase reporter activation was measured with the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions.
Chromosome breaks analysis
HeLa cells were transduced and treated with mitomycin C (MMC; 100 nM) for 24 hours, followed by 3 hours of treatment with colcemid (200 ng/mL). The cells were collected and swollen in hypotonic buffer (75 mM KCl) for 20 minutes at 37°C, fixed for 1 hour on ice using a 3:1 ratio of methanol:acetic acid, and dropped onto slides. The slides were stained with 5% Gurr’s KaryoMAX Giemsa stain (Gibco, Carlsbad, CA).
Immunofluorescence
For FA proteins and CtBP1 localization, HeLa cells were fixed in methanol:acetone (3:7 v/v) and permeabilized with 0.1% saponin before immunofluorescent staining. For FANCD2 foci analysis, HeLa-transduced cells were treated with MMC (150 nM) or hydroxyurea (HU; 2 mM) for 16 hours. Cells were fixed in 4% paraformaldehyde and permeabilized in 0.3% Triton X-100 before immunofluorescent staining. The slides were mounted with DAPI-Fluoromount-G (Southern Biotech, Birmingham, AL)
RNA isolation, first strand cDNA synthesis, and quantitative PCR
HeLa cells were transduced with lentiviral particles as described above. Total RNA was extracted with Trizol or the RNeasy Mini Kit with DNAse digestion (Qiagen, Valencia, CA) and resuspended in PCR-grade water. To ensure adequate quality, RNA was analyzed with an Agilent Bioanalyser (Agilent Technologies, Santa Clara, CA) in the Quebec Genomics Center Microarray Core Facility. First-strand cDNA syntheses were performed using the iScript cDNA synthesis system (Bio-Rad, Mississauga, ON, Canada) according to the manufacturer’s instructions. PCR reactions were monitored in real time with a LightCycler System (Roche Applied Science, Indianapolis, IN) and SsoFast EvaGreen Supermix (Bio-Rad). The primer pairs used and quantitative PCR conditions are summarized in Materials and Methods. Data were analyzed with LightCycler Software version 3.5 (Roche Applied Science).
Microarray analyses
Total RNA extracts from 3 different samples of each scrambled CtBPs and FANCD2 shRNA-treated HeLa cells were subjected to gene expression profiling via microarray analysis. Gene expression profiles were determined with Affymetrix GeneChip Human Gene 1.0 ST Arrays (28 869 probes sets) at the Quebec Genomics Center Microarray Core Facility (GEO accession number: GSE43330). Genes exhibiting differential upregulation and downregulation patterns were selected on the basis of a >2-fold change with greater than 95% confidence relative to control cells expressing scrambled shRNA.
DKK1 ELISA
Supernatant from HeLa transduced cell lines in culture and peripheral blood from 10-month-old wild-type FancA−/− and FancC−/− mice were collected and subjected to an enzyme-linked immunosorbent assay (ELISA) to evaluate DKK1 concentration by using DKK100 and MKK100 ELISA kits (R&D Systems, Minneapolis, MN), according to the manufacturer’s instructions. All animal procedures were performed according to protocols approved by the Animal Care Committee of Laval University.
Statistical analyses
Data were expressed as the means ± standard error of the mean. Statistical analyses were performed with GraphPad Prism software (version 5.0b; GraphPad Software, San Diego, CA), and the tests used included one-way analysis of variance and 2-tailed Student t tests. P values less than .05 were considered significant.
Results
CtBP1 directly interacts with FANCC
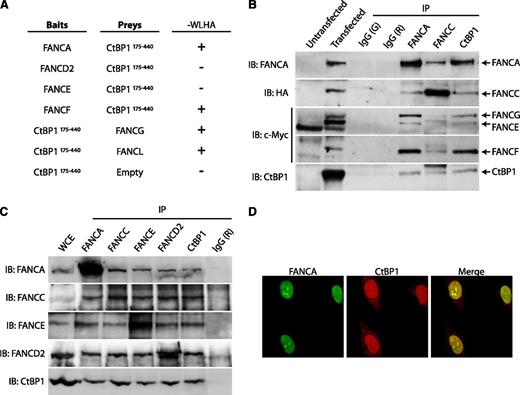
On the basis of earlier observations that FANCC undergoes proteolytic modification,16 we sought to determine the biological activity of the resultant FANCC fragments. Because protein–protein interactions help identify protein functions, we mimicked FANCC cleavage products with N-terminal (FANCC1-306) and C-terminal (FANCC307-558) FANCC truncations, which were used as bait proteins for yeast 2-hybrid analyses. Three independent screens were performed in which 20 positive yeast colonies were obtained. Of the positive plasmids isolated from these colonies, one was FANCC and another corresponded to the FANCG-interacting protein peroxyredoxin 3.25 Three of the positive clones encoded the C-terminal portion (aa 175-440) of CtBP1 (Figure 1A). This CtBP1175-440 clone was retested in yeast 2-hybrid screens against both N- and C-terminal fragments, in addition to full-length FANCC. The results revealed that CtBP1175-440 interacts directly with N-terminal FANCC (FANCC1-306) but not with C-terminal FANCC (FANCC307-558; Figure 1B). As expected, no interaction was detected between CtBP1175-440 and FANCC307-558 harboring the L554P mutation (data not shown). Surprisingly, no interaction was detected between CtBP1175-440 and full-length FANCC, suggesting that in yeasts, the conformation of full-length FANCC may prevent its interaction with CtBP1 or that the full-length FANCC cannot enter the yeast nucleus required to activate reporter genes. Another possibility is that because the CtBP1175-440 clone is missing part of the RRT-binding groove and part of the dimerization interface,26,27 these protein domains may be required for a stable interaction with full-length FANCC.
CtBP1 interacts with the N-terminal region of FANCC in yeast. (A) Schematic representation of the mammalian protein CtBP1. Two isoforms (CtBP1-L and CtBP1-S) are generated by two mRNA splice variants. The various CtBP1 domains are indicated; these include a D-isomer–specific 2-hydroxy acid dehydrogenase domain (D2-HDH), PLDLS-binding sites, a dimerization domain, and an RRT-binding groove. Phosphorylation sites are shown as circles. The CtBP1 clone obtained from yeast screening corresponds to amino acids 175-440. (B) Yeast 2-hybrid assay with FANCC and CtBP1175-440 proteins. The AH109 yeast strain was co-transformed with FANCC constructs expressing full-length or truncated FANCC as baits and with CtBP1175-440 as prey and was assayed for interaction by plating with dropout medium lacking tryptophan, leucine, histidine, and adenine (−WLHA). Negative and positive interactions are indicated as (−) and (+), respectively. The negative controls include CtBP1175-440 cotransformed with the empty bait vector. Positive controls include p53 bait and SV40 T antigen prey vectors (not shown). Each experiment was performed a minimum of 3 times in triplicate with each gene cloned into either the bait or prey vector.
CtBP1 interacts with the N-terminal region of FANCC in yeast. (A) Schematic representation of the mammalian protein CtBP1. Two isoforms (CtBP1-L and CtBP1-S) are generated by two mRNA splice variants. The various CtBP1 domains are indicated; these include a D-isomer–specific 2-hydroxy acid dehydrogenase domain (D2-HDH), PLDLS-binding sites, a dimerization domain, and an RRT-binding groove. Phosphorylation sites are shown as circles. The CtBP1 clone obtained from yeast screening corresponds to amino acids 175-440. (B) Yeast 2-hybrid assay with FANCC and CtBP1175-440 proteins. The AH109 yeast strain was co-transformed with FANCC constructs expressing full-length or truncated FANCC as baits and with CtBP1175-440 as prey and was assayed for interaction by plating with dropout medium lacking tryptophan, leucine, histidine, and adenine (−WLHA). Negative and positive interactions are indicated as (−) and (+), respectively. The negative controls include CtBP1175-440 cotransformed with the empty bait vector. Positive controls include p53 bait and SV40 T antigen prey vectors (not shown). Each experiment was performed a minimum of 3 times in triplicate with each gene cloned into either the bait or prey vector.
To determine whether full-length CtBP1 interacts with full-length FANCC in human cells, immunoprecipitation studies were performed in HEK293T cells transiently expressing various FANCC constructs, including full-length FANCC, FANCC1-306, FANCC307-558, and the naturally occurring amino terminal–truncated FANCC methionine55 polypeptide (FANCC55-558).28 FANCC55-558 mimics the 50-kDa amino terminal–truncated FANCC polypeptide resulting from reinitiation of translation at methionine 55 occurring in cells from patients with a 322delG mutation. Cells overexpressing GFP–tagged FANCC constructs (GFP-FANCC8-558, GFP-FANCC55-558, GFP-FANCC1-306, or GFP-FANCC307-558) were subjected to co-immunoprecipitation with anti-GFP or anti-CtBP1 antibodies. Western blotting confirmed that endogenous full-length CtBP1 interacts with all FANCC constructs tested, including FANCC1-306, FANCC307-558, FANCC55-558, and full-length FANCC (Figure 2A). These results suggest that full-length CtBP1 interacts with full-length FANCC as well as with FANCC fragments. The fact that CtBP1 co-immunoprecipitates with FANCC55-558 suggests that CtBP1-FANCC interaction is conserved in patients with 322delG mutation.
CtBP1 interacts with FANCC in cells. (A) Co-immunoprecipitation of CtBP1 with FANCC. HEK293T cells were transfected with GFP-tagged FANCC constructs expressing full-length (FANCC8-558) or truncated FANCC (FANCC 55-558, FANCC1-306, and FANCC307-558) constructs. Whole cell extracts (WCEs) were subjected to immunoprecipitation (IP) with antibodies against GFP or CtBP1. Negative IP controls were performed using mouse immunoglobulin G (IgG) (M). IPs were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted (IB) with the indicated antibodies. Data shown are representative of at least 3 independent experiments. (B) Co-IP of endogenous FANCC and CtBP1 proteins from HEK293T cell extracts using anti-CtBP1 or anti-FANCC antibodies. Negative IP control was performed using rabbit IgG (R). Shown is 1 of 5 representative experiments. (C) CtBP1 and FANCC localization in HeLa cells. Cells were double-stained with anti-CtBP1 (green) and anti-FANCC (red) antibodies and were visualized via confocal fluorescence microscopy using a Nikon E800 microscope equipped with a C1 confocal system at 100× magnification. White boxes in the left panel indicate selected cells that have been magnified in the right panel. Data shown are representative of 3 experiments in which at least 25 cells were analyzed.
CtBP1 interacts with FANCC in cells. (A) Co-immunoprecipitation of CtBP1 with FANCC. HEK293T cells were transfected with GFP-tagged FANCC constructs expressing full-length (FANCC8-558) or truncated FANCC (FANCC 55-558, FANCC1-306, and FANCC307-558) constructs. Whole cell extracts (WCEs) were subjected to immunoprecipitation (IP) with antibodies against GFP or CtBP1. Negative IP controls were performed using mouse immunoglobulin G (IgG) (M). IPs were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted (IB) with the indicated antibodies. Data shown are representative of at least 3 independent experiments. (B) Co-IP of endogenous FANCC and CtBP1 proteins from HEK293T cell extracts using anti-CtBP1 or anti-FANCC antibodies. Negative IP control was performed using rabbit IgG (R). Shown is 1 of 5 representative experiments. (C) CtBP1 and FANCC localization in HeLa cells. Cells were double-stained with anti-CtBP1 (green) and anti-FANCC (red) antibodies and were visualized via confocal fluorescence microscopy using a Nikon E800 microscope equipped with a C1 confocal system at 100× magnification. White boxes in the left panel indicate selected cells that have been magnified in the right panel. Data shown are representative of 3 experiments in which at least 25 cells were analyzed.
In addition, western blot analyses of immunoprecipitates from endogenous proteins with anti-FANCC or anti-CtBP1 antibodies revealed that both endogenous FANCC and CtBP1 proteins were co-immunoprecipitated, confirming that CtBP1 interacts with FANCC in cells (Figure 2B). As expected, confocal immunofluorescence microscopy of endogenous proteins revealed that CtBP1 localized with FANCC in the same cellular compartment, particularly in the nucleus (Figure 2C). Collectively, our data show that CtBP1 is a novel interacting partner of FANCC.
CtBP1 interacts with other components of the FA core complex
Because nuclear FANCC is a component of the FA core complex, we next examined potential interactions between CtBP1 and other FA core complex proteins. First, we used the yeast 2-hybrid system to test for direct interactions between FA core complex components and the C-terminal fragment of CtBP1, CtBP1175-440. The results of this analysis revealed that CtBP1175-440 interacts directly with FANCA, FANCF, FANCG, and FANCL but does not interact directly with FANCD2 or FANCE (Figure 3A).
CtBP1 interacts with components of the FA core complex. (A) CtBP1 interacts with FANCA, FANCF, FANCG and FANCL in yeasts. Yeast 2-hybrid assays were performed using FA core complex components as baits and CtBP1175-440 as prey. Negative and positive interactions are indicated as (−) and (+), respectively. The negative control consisted of a CtBP1175-440 bait vector with an empty prey vector. Positive controls included p53 bait and SV40 T antigen prey vectors (not shown). Each experiment was performed a minimum of 3 times in triplicate with each gene cloned into either the bait or prey vector (except for FANCG and FANCL). (B) CtBP1 co-immunoprecipitates with FA core complex proteins. HEK293T cells were cotransfected with T7-tagged CtBP1 and plasmids expressing FANCA, FANCC, FANCE, FANCF, FANCG, or FANCL. WCEs were then subjected to IP, followed by SDS-PAGE and IB with the indicated antibodies. Negative IP controls were performed using goat (G) and rabbit (R) IgG. Data shown are representative of 2 identical and separate experiments. (C) Co-IP of endogenous proteins from FANCD2-complemented PD20 cell extracts with anti-CtBP1, anti-FANCA, anti-FANCC, anti-FANCE or anti-FANCD2 antibodies. Negative IP control was done using rabbit IgG (R). Results shown are representative of at least 4 independent experiments performed in different cell lines, including HEK293T, HeLa, FANCA-complemented PD220, or FANCD2-complemented PD20 cells. (D) Confocal immunofluorescence of CtBP1 and FANCA localization in HeLa cells. Cells were double-stained with anti-FANCA (green) and anti-CtBP1 (red) antibodies and were visualized using a Nikon E800 microscope equipped with C1 confocal system at 100× magnification. Shown are 3 representative experiments in which at least 25 cells were analyzed per experiment.
CtBP1 interacts with components of the FA core complex. (A) CtBP1 interacts with FANCA, FANCF, FANCG and FANCL in yeasts. Yeast 2-hybrid assays were performed using FA core complex components as baits and CtBP1175-440 as prey. Negative and positive interactions are indicated as (−) and (+), respectively. The negative control consisted of a CtBP1175-440 bait vector with an empty prey vector. Positive controls included p53 bait and SV40 T antigen prey vectors (not shown). Each experiment was performed a minimum of 3 times in triplicate with each gene cloned into either the bait or prey vector (except for FANCG and FANCL). (B) CtBP1 co-immunoprecipitates with FA core complex proteins. HEK293T cells were cotransfected with T7-tagged CtBP1 and plasmids expressing FANCA, FANCC, FANCE, FANCF, FANCG, or FANCL. WCEs were then subjected to IP, followed by SDS-PAGE and IB with the indicated antibodies. Negative IP controls were performed using goat (G) and rabbit (R) IgG. Data shown are representative of 2 identical and separate experiments. (C) Co-IP of endogenous proteins from FANCD2-complemented PD20 cell extracts with anti-CtBP1, anti-FANCA, anti-FANCC, anti-FANCE or anti-FANCD2 antibodies. Negative IP control was done using rabbit IgG (R). Results shown are representative of at least 4 independent experiments performed in different cell lines, including HEK293T, HeLa, FANCA-complemented PD220, or FANCD2-complemented PD20 cells. (D) Confocal immunofluorescence of CtBP1 and FANCA localization in HeLa cells. Cells were double-stained with anti-FANCA (green) and anti-CtBP1 (red) antibodies and were visualized using a Nikon E800 microscope equipped with C1 confocal system at 100× magnification. Shown are 3 representative experiments in which at least 25 cells were analyzed per experiment.
To confirm these interactions in cells, co-immunoprecipitation studies were performed in human cells transfected with FA core complex components both separately and in conjunction with CtBP1. For each FA protein expressed with CtBP1, immunoprecipitations were performed with antibodies against the epitope tag. As expected, each FA protein that was expressed with CtBP1 was found in immunoprecipitates with CtBP1, including FANCA, FANCC, FANCE, FANCF, FANCG, and FANCL. No proteins were detected in immunoglobulin G control immunoprecipitates (supplemental Figure 2). On the basis of these results, we next determined whether CtBP1 interacted with the FA core complex. Co-immunoprecipitation experiments were performed with anti-FANCA, anti-FANCC, or anti-CtBP1 antibodies in cells co-expressing FANCA, FANCC, FANCE, FANCF, FANCG, FANCL, and CtBP1. Immunoprecipitates obtained with antibodies against FANCA, FANCC, and CtBP1, but not control immunoglobulin G, contained CtBP1 and all core complex components tested, including FANCA, FANCC, FANCE, FANCF, and FANCG (Figure 3B). These results confirmed the yeast 2-hybrid findings and suggested that an interaction occurs between CtBP1 and several FA core complex components.
To confirm that interaction of CtBP1 with FA core complex proteins is not a consequence of overexpression, endogenous proteins were immunoprecipitated with anti-FANCA, anti-FANCC, anti-FANCE, anti-FANCD2, or anti-CtBP1 antibodies. Western blot analysis of the immunoprecipitates obtained revealed that FANCA, FANCC, FANCE, and FANCD2 co-immunoprecipitated together with CtBP1 (Figure 3C). These results confirm that CtBP1 interacts with endogenous FA proteins, including FANCA, FANCC, FANCE, and FANCD2 and suggest that CtBP1 interacts with the FA core complex as well as the core complex substrate FANCD2.
Next, we examined the cellular localization of CtBP1 and FANCA as a representative core complex protein. In agreement with the immunoprecipitation results, confocal microscopy of endogenous CtBP1 and FANCA proteins revealed that these 2 proteins are located together in the nucleus (Figure 3D). Collectively, our data reveal that CtBP1 is a novel interacting partner of FA core complex proteins.
CtBP1 is not essential to the integrity or the activity of the FA core complex
Each of the FA core complex proteins has been shown to be essential to the integrity and nuclear localization of the core complex, which reflects the interdependence of FA core complex proteins. To determine whether CtBP1 is essential to the integrity of the core complex, immunoprecipitation and immunofluorescence analyses were performed with CtBP1-depleted cells (CtBP1i). Immunoprecipitations with anti-FANCA, anti-FANCE, anti-FANCD2, or anti-CtBP1 antibodies were performed. Western blots of CtBP1i immunoprecipitates indicated that depletion of CtBP1 did not affect the interaction between FA core complex proteins (data not shown). Because both CtBP1 and its homolog CtBP2 act as homodimers or heterodimers, are functionally redundant, and have overlapping roles,21,26 we also tested the integrity of the FA core complex in CtBP1-/CtBP2-depleted cells (CtBPi). The results from this latter experiment reaffirmed that CtBP1 is not required for FA core complex integrity and suggest that it may act downstream of the FA core complex formation (Figure 4A).
CtBP1 is not essential for the stability or the activity of the FA core complex. (A) Functional FA core complex assembly in cells depleted of CtBP. Co-immunoprecipitations were performed in HEK293T cells transduced with shRNA against CtBP1/CtBP2 (CtBPi) or with shRNA against scrambled sequences (control). Protein extracts were collected 3 days post-transductions and were subjected to IP with anti-FANCA, anti-FANCE, anti-FANCD2, and anti-CtBP1 antibodies. The IPs were resolved by SDS-PAGE and analyzed via IB with the indicated antibodies. Negative IP control was performed using rabbit IgG (R). Results shown are representative of 2 independent experiments. (B) CtBP1 is not required for FA core complex activity. HeLa cells transduced with shRNA against CtBP1 (CtBP1i), CtBP2 (CtBP2i), CtBP1/CtBP2 (CtBPi), FANCA (FANCAi) or FANCA/CtBP1/CtBP2 (CtBPi/FANCAi) were treated with 300 nM MMC or 2 mM HU for 16 hours. Equal amounts of protein were subjected to IB analysis. The FANCD2-Ub:FANCD2 ratios (L:S) are indicated below each sample. Controls included untreated cells (not shown) and cells transduced with shRNA against scrambled sequences (Control). Results shown are representative of 5 separate experiments. (C) CtBP1 is not required for FANCD2 foci formation. CtBP1- (CtBP1i) and FANCA- (FANCAi) depleted HeLa cells were treated with 150 nM MMC or 2 mM HU and subjected to immunofluorescence (IF) analysis and visualized using a Nikon E800 microscope equipped with C1 confocal system (100× magnification). White boxes in the left panels indicate selected cells magnified in the right panel for each cell line. (D) FANCD2 foci formation in CtBP-depleted cells. HeLa cells depleted of CtBP1 and/or CtBP2 (CtBP1i, CtBP2i, CtBPi), FANCA (FANCAi) or CtBP1/CtBP2 and FANCA (CtBPi/FANCAi) were treated as in panel C. Cells containing more than 10 bright foci were scored as FANCD2 foci-positive cells. Fluorescence intensity settings were determined using untreated cells. A minimum of 150 cells was scored for each condition. Values are expressed as the mean percentages of FANCD2 foci positive cells ± standard error of the mean (SEM). Ctl, control. ***P ≤ .001.
CtBP1 is not essential for the stability or the activity of the FA core complex. (A) Functional FA core complex assembly in cells depleted of CtBP. Co-immunoprecipitations were performed in HEK293T cells transduced with shRNA against CtBP1/CtBP2 (CtBPi) or with shRNA against scrambled sequences (control). Protein extracts were collected 3 days post-transductions and were subjected to IP with anti-FANCA, anti-FANCE, anti-FANCD2, and anti-CtBP1 antibodies. The IPs were resolved by SDS-PAGE and analyzed via IB with the indicated antibodies. Negative IP control was performed using rabbit IgG (R). Results shown are representative of 2 independent experiments. (B) CtBP1 is not required for FA core complex activity. HeLa cells transduced with shRNA against CtBP1 (CtBP1i), CtBP2 (CtBP2i), CtBP1/CtBP2 (CtBPi), FANCA (FANCAi) or FANCA/CtBP1/CtBP2 (CtBPi/FANCAi) were treated with 300 nM MMC or 2 mM HU for 16 hours. Equal amounts of protein were subjected to IB analysis. The FANCD2-Ub:FANCD2 ratios (L:S) are indicated below each sample. Controls included untreated cells (not shown) and cells transduced with shRNA against scrambled sequences (Control). Results shown are representative of 5 separate experiments. (C) CtBP1 is not required for FANCD2 foci formation. CtBP1- (CtBP1i) and FANCA- (FANCAi) depleted HeLa cells were treated with 150 nM MMC or 2 mM HU and subjected to immunofluorescence (IF) analysis and visualized using a Nikon E800 microscope equipped with C1 confocal system (100× magnification). White boxes in the left panels indicate selected cells magnified in the right panel for each cell line. (D) FANCD2 foci formation in CtBP-depleted cells. HeLa cells depleted of CtBP1 and/or CtBP2 (CtBP1i, CtBP2i, CtBPi), FANCA (FANCAi) or CtBP1/CtBP2 and FANCA (CtBPi/FANCAi) were treated as in panel C. Cells containing more than 10 bright foci were scored as FANCD2 foci-positive cells. Fluorescence intensity settings were determined using untreated cells. A minimum of 150 cells was scored for each condition. Values are expressed as the mean percentages of FANCD2 foci positive cells ± standard error of the mean (SEM). Ctl, control. ***P ≤ .001.
The FA pathway is generally considered to be a canonical pathway; the primary function of the FA core complex is to monoubiquitinate two downstream substrates, FANCD2 and FANCI, in response to DNA crosslinks. Mutations in any FA core complex proteins compromise this activity, which is reflected by a reduction in FANCD2 and FANCI monoubiquitination.29,30 In addition, FANCD2 monoubiquitination is both necessary and sufficient to induce FANCD2 relocalization to nuclear foci and is therefore an indicator of intact FA core complex activity. To determine whether CtBP1 influences this FA core complex activity, we examined the monoubiquitination and relocalization of FANCD2 to nuclear foci following DNA damage in CtBP1i cells relative to FANCAi cells (which are FA core complex deficient). We also evaluated the activity of the FA core complex in CtBP2i and CtBPi cells to exclude overlapping roles between CtBP1 and CtBP2 and in CtBPi/FANCAi cells to verify the possibility of epistatic effects between CtBP1 and the FA core complex. Transduced cells were treated with MMC or HU for 16 hours prior to protein extraction. Extracts were examined for FANCD2 monoubiquitination (FANCD2-Ub), and the FANCD2-Ub:FANCD2 ratio was evaluated following western blotting. The results indicated that, upon MMC treatment, FANCD2-Ub was strongly upregulated in CtBP1i cells in a manner that was similar to that in control cells (Figure 4B). As expected, FANCAi cells exhibited greatly reduced levels of FANCD2-Ub (ie, 2.5 times lower than controls). Depletion of CtBP1, its homolog CtBP2, or CtBP1 and CtBP2 together did not affect the accumulation of FANCD2-Ub in response to DNA damage. Depletion of both CtBP homologs in FANCAi cells did not alter the reduction in FANCD2-Ub upon treatment with MMC or HU. These results indicate that CtBP1 is not required for FA core complex ubiquitin ligase activity. In addition, CtBP1i cells exhibited no change in the formation of nuclear FANCD2 foci after DNA crosslink damage, which correlated with FANCD2 monoubiquitination levels determined by western blot (Figure 4C). Quantification of FANCD2 nuclear foci–positive cells after treatment revealed no difference between CtBP1i and control cells in the percentage of cells with 10 or more foci; in contrast, the expected reduction in the number of cells with FANCD2 foci was observed in FANCAi or CtBPi/FANCAi cells (Figure 4D). These results suggest that CtBP1 is not required for the FA core complex assembly and activity in its response to DNA crosslink damage. These results also suggest that CtBP1 together with FA proteins may act downstream or independently of the FANCD2 signal.
CtBP1 is essential for cell survival and chromosomal integrity
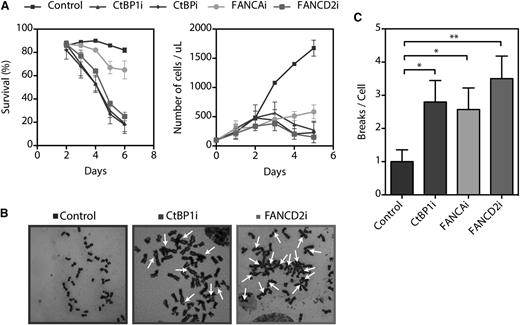
To understand the functional mechanism of the interaction between CtBP1 and FA proteins, we attempted to establish stable CtBP1i cell lines. However, no stable CtBP1i cell lines could be established because of reduced cell growth and increased cell death (Figure 5A and supplemental Figure 3). CtBP1i as well as CtBPi cells did not survive beyond 6 days of culture, suggesting that CtBP1 and its homolog CtBP2 are essential for cell survival in cultured cells, as previously reported.31 Thus, we performed cell viability assays using an acetomethoxy derivate of calcein and showed that CtBP1i, CtBPi, and FANCD2i cells began to die at 3 days post-transduction (Figure 5A, left graph). Cellular death increased markedly at day 4, and all remaining cells died at 7 days. While FANCAi cells survived up to 7 days post-transduction, their rate of proliferation relative to scrambled shRNA control cells was dramatically reduced (Figure 5A, right graph).
CtBP1 is essential for survival and proliferation. (A) Survival and proliferation curves of HeLa cells transduced with shRNA against CtBP1 (CtBP1i), CtBP1/CtBP2 (CtBPi), FANCA (FANCAi), or FANCD2 (FANCD2i). The percentages of living cells were scored via calcein-AM staining on each day post-transduction. Living cells were scored with a FACSCalibur cytometer (BD Biosciences) and the percentage of live cells in each sample was determined with CellQuest Pro software (BD Biosciences, Mississauga, ON). (B) Representative metaphase spread of HeLa cells transduced with shRNA against CtBP1 (CtBP1i), FANCA (FANCAi), or FANCD2 (FANCD2i) following a 16 hours exposure to 150 nM MMC. Arrows indicate chromatid-type breakage and radial rejoining. (C) Numbers of chromosomal breaks and aberrations per metaphase nucleus identified in CtBP1-depleted (CtBP1i), FANCA-depleted (FANCAi), and FANCD2-depleted (FANCD2i) HeLa cells. Each experiment was performed a minimum of 3 times. To evaluate chromosomal abnormalities, 25 metaphase nuclei were analyzed per experiment with a Carl Zeiss Axio Imager M2 microscope equipped with an Axiocam MRm camera and Axiovision Rel.4.8 software (100× magnification). Values represent the means ± SEM. *P ≤ .05; **P ≤ .01 vs control shRNA scrambled transduced cells.
CtBP1 is essential for survival and proliferation. (A) Survival and proliferation curves of HeLa cells transduced with shRNA against CtBP1 (CtBP1i), CtBP1/CtBP2 (CtBPi), FANCA (FANCAi), or FANCD2 (FANCD2i). The percentages of living cells were scored via calcein-AM staining on each day post-transduction. Living cells were scored with a FACSCalibur cytometer (BD Biosciences) and the percentage of live cells in each sample was determined with CellQuest Pro software (BD Biosciences, Mississauga, ON). (B) Representative metaphase spread of HeLa cells transduced with shRNA against CtBP1 (CtBP1i), FANCA (FANCAi), or FANCD2 (FANCD2i) following a 16 hours exposure to 150 nM MMC. Arrows indicate chromatid-type breakage and radial rejoining. (C) Numbers of chromosomal breaks and aberrations per metaphase nucleus identified in CtBP1-depleted (CtBP1i), FANCA-depleted (FANCAi), and FANCD2-depleted (FANCD2i) HeLa cells. Each experiment was performed a minimum of 3 times. To evaluate chromosomal abnormalities, 25 metaphase nuclei were analyzed per experiment with a Carl Zeiss Axio Imager M2 microscope equipped with an Axiocam MRm camera and Axiovision Rel.4.8 software (100× magnification). Values represent the means ± SEM. *P ≤ .05; **P ≤ .01 vs control shRNA scrambled transduced cells.
We next evaluated the cell cycle status of CtBP1i, FANCAi, and FANCD2i cells by flow cytometry using propidium iodide DNA-binding dye. The proportion of CtBP1i and FANCD2i cells in both G1-S and G2-M phases of the cell cycle decreased progressively during culture relative to control cells, which indicated that there was no delay in the G2-M phase transition (supplemental Figure 3A). The decreased number of cells in both G1-S and G2-M phases of the cell cycle is associated with an increase in the number of cells in sub-G1, which is indicative of apoptotic death. In fact, western blot analyses revealed the appearance of cleaved poly(ADP-ribose) polymerase and activated caspase-3 (supplemental Figure 3B). Additional subtle changes in cell cycle phases were observed in FANCAi cells, for which a reduced number of G1-S and G2-M cells were associated with increased polyploidy (4N).
Because CtBP1i cells progressively died of apoptosis, clonogenic and survival assays to establish their sensitivity to DNA crosslinking agents could not be performed. However, chromosomal breakage analysis after treatment with MMC and diepoxybutane, which is the standard method for FA testing, was evaluated in CtBP1i, FANCAi, and FANCD2i cells. Chromosomal breakage analysis revealed that depletion of CtBP1 causes extensive chromosomal aberrations in response to MMC treatments, similar to what was observed in FANCD2i cells, which are representative of FA cells (Figure 5B). Quantification of total chromosomal damages, including rearrangement, breakage and radial formation per cell, revealed significant differences between CtBP1i cells and control cells that were similar to those observed in cells with depleted FANCA or FANCD2 (Figure 5C). These results suggest that CtBP1 is essential for cell proliferation, cell survival, and chromosome stability. This is consistent with previous reports showing that CtBP1 is involved in mitotic fidelity, including chromosome pairing during cellular division.18,31 Collectively, our results suggest that CtBP1-depleted cells lead to defects that characterize cells from FA patients.
Depletion of CtBP1 and FANCD2 modulate DKK1 expression
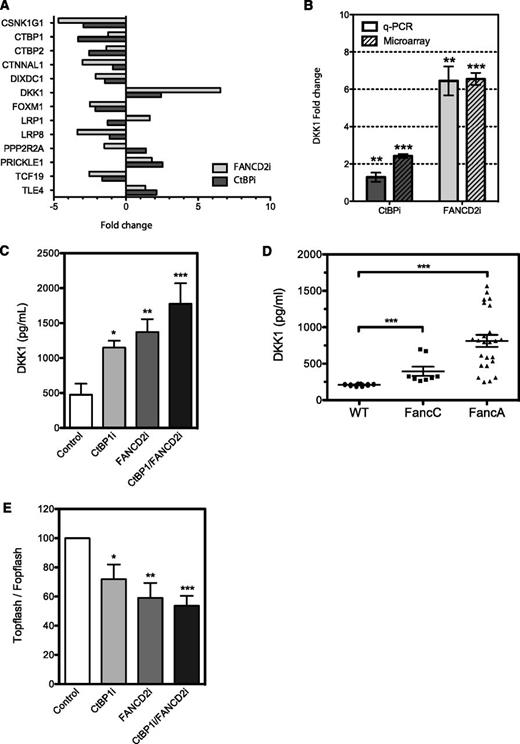
Because the primary role of CtBP1 is to negatively regulate transcription of Wnt response genes and because both CtBP- and FANCD2-depleted cells exhibited similar growth and proliferation kinetics, we investigated the gene expression profiles of these cells. RNA from CtBPi and FANCD2i cells from three independent transduction experiments were extracted and processed for microarray analysis. Differentially expressed genes were defined as those with at least a 2-fold change in expression relative to control cells. The various comparisons included the identification of genes differentially expressed between samples from CtBPi, FANCD2i, and controls. Of the 230 genes in CtBPi cells and 737 genes in FANCD2i cells that were up- or downregulated, 45 genes were common in both cell lines (supplemental Figure 4). As expected, CtBPi cells exhibited downregulation of both CtBP1 and CtBP2 homologs. Ingenuity system pathway analysis revealed that the Wnt pathway was dysregulated in CtBPi but also in FANCD2i cells. Genes involved in Wnt activation were downegulated, including the casein kinase 1 isoform gamma 1 (CSNK1G1), which is involved in phosphorylation of the Wnt relay protein Disheveled, the catenin alpha-like 1 (CTNNAL1), the DIX domain–containing 1 gene (DIXDC1), the transcriptional activator forkhead box M1 (FOXM1), the low-density lipoprotein receptor–related protein 1 and 8 (LRP1 and LRP8), and the transcription factor TCF19, whereas genes involved in negative regulation of the Wnt signal were found upregulated, including the potent secreted Wnt antagonist DKK1, the Wnt/planar polarity pathway protein Prickle 1 (PRKL-1), and the corepressor transducing-like enhancer of split 4 (TLE4) (Figure 6A). Surprisingly, one of the most upregulated genes in FANCD2i cells was DKK1, which exhibited a 6.5-fold upregulation.
CtBP1 and FANCD2 regulate DKK1 expression. (A) Up- and downregulation of genes involved in Wnt pathways found in HeLa cells depleted of CtBP1/CtBP2 (CtBPi) or FANCD2 (FANCD2i) from microarray data. Data represent fold change in mRNA expression relative to control cells expressing scrambled shRNA. (B) DKK1 expression analysis in HeLa cells transduced with shRNA against CtBP1/CtBP2 (CtBPi) or FANCD2 (FANCD2i). DKK1 mRNA expression was quantified via quantitative PCR (q-PCR) and normalized to expression of the housekeeping gene HPRT1. Data are expressed as the mean fold changes ± SEM relative to control cells expressing scrambled shRNA from 4 different experiments performed in duplicate. (C) DKK1 levels found in HeLa cells depleted of CtBP1 (CtBP1i), FANCD2 (FANCD2i), or both (CtBP1/FANCD2i). DKK1 protein levels in cell culture media were determined using a DKK1 detection ELISA kit and compared with control cells expressing scrambled shRNA (Control). Results are shown as the mean DKK1 concentration ± SEM of 3 independent experiments performed in duplicate, normalized to cell numbers and culture media volumes. (D) Serum samples from 10-month-old wild-type (WT; n = 7), FancA−/− (n = 10), and FancC−/− (n = 4) mouse littermates were collected, and Dkk1 plasma levels were measured in duplicates using a Dkk1 detection ELISA kit. (E) Luciferase reporter assays performed in HeLa cells depleted of CtBP1, FANCD2 or CtBP1/FANCD2 following transfection of the pTOPflash TCF/LEF reporter construct or the pFOPflash TCF/LEF mutant construct. Results were compared with control cells expressing scrambled shRNA. Results are expressed as percent TOPflash/FOPflash activity of control cells from 4 different experiments performed in duplicate. * P ≤ .05, **P ≤ .01, *** P ≤ .001.
CtBP1 and FANCD2 regulate DKK1 expression. (A) Up- and downregulation of genes involved in Wnt pathways found in HeLa cells depleted of CtBP1/CtBP2 (CtBPi) or FANCD2 (FANCD2i) from microarray data. Data represent fold change in mRNA expression relative to control cells expressing scrambled shRNA. (B) DKK1 expression analysis in HeLa cells transduced with shRNA against CtBP1/CtBP2 (CtBPi) or FANCD2 (FANCD2i). DKK1 mRNA expression was quantified via quantitative PCR (q-PCR) and normalized to expression of the housekeeping gene HPRT1. Data are expressed as the mean fold changes ± SEM relative to control cells expressing scrambled shRNA from 4 different experiments performed in duplicate. (C) DKK1 levels found in HeLa cells depleted of CtBP1 (CtBP1i), FANCD2 (FANCD2i), or both (CtBP1/FANCD2i). DKK1 protein levels in cell culture media were determined using a DKK1 detection ELISA kit and compared with control cells expressing scrambled shRNA (Control). Results are shown as the mean DKK1 concentration ± SEM of 3 independent experiments performed in duplicate, normalized to cell numbers and culture media volumes. (D) Serum samples from 10-month-old wild-type (WT; n = 7), FancA−/− (n = 10), and FancC−/− (n = 4) mouse littermates were collected, and Dkk1 plasma levels were measured in duplicates using a Dkk1 detection ELISA kit. (E) Luciferase reporter assays performed in HeLa cells depleted of CtBP1, FANCD2 or CtBP1/FANCD2 following transfection of the pTOPflash TCF/LEF reporter construct or the pFOPflash TCF/LEF mutant construct. Results were compared with control cells expressing scrambled shRNA. Results are expressed as percent TOPflash/FOPflash activity of control cells from 4 different experiments performed in duplicate. * P ≤ .05, **P ≤ .01, *** P ≤ .001.
To confirm the microarray findings, CtBPi and FANCD2i cells were freshly prepared and tested for DKK1 messenger RNA (mRNA) and protein expression. Upregulation of DKK1 mRNA expression was identified in both CtBPi and FANCD2i cells, with a more dramatic effect in FANCD2i cells that confirmed the expression profiling results (Figure 6B). This upregulation in DKK1 mRNA translates into an increase in protein expression and secretion in both CtBP1i and FANCD2i cells, which were measured using ELISA detection of DKK1 on supernatant from these cells (Figure 6C). Depletion of both CtBP1 and FANCD2 lead to a further increase in DKK1 protein expression. To determine whether the increase in DKK1 expression and secretion was not limited to cultured cells, we measured Dkk1 protein levels in sera from FancA and FancC knockout mice by using an ELISA detection assay. Results show that the increase in DKK1 protein expression observed in cells is paralleled by elevated Dkk1 levels detected in sera from both FancA−/− and FancC−/− knockout mice compared with wild-type littermates (Figure 6D). Because DKK1 is an antagonist of the Wnt signal and the DKK1 gene is a target of TCF/β-catenin, we sought to determine whether elevated levels of DKK1 translate into changes in TCF/β-catenin activity. CtBP1i, FANCD2i, and CtBP1/FANCD2i cells were transfected with the TCF/β-catenin reporter construct TOPflash or the negative FOPflash vector and tested for reporter activation. Results show that CtBP1i and FANCD2i cells had reduced TCF/LEF reporter activation and that cells depleted in both CtBP1 and FANCD2 showed further reduction in TCF/LEF activity (50% reduction in TOPflash activation; Figure 6E). These results indicate that overproduction of DKK1 has a negative impact on TCF/β-catenin activity. Altogether, our results suggest that FA and CtBP1 proteins play a role in the regulation of the Wnt antagonist DKK1.
Discussion
The aim of our study was to characterize the biological function of FANCC by identifying novel interacting partners that could provide clues to molecular events leading to bone marrow failure. Our data provide evidence of interplay between the FA pathway and the Wnt pathway via CtBP1, a protein that is primarily involved in transcriptional regulation of the Wnt signal. We found that CtBP1 interacts with FANCC and several components of the FA core complex. Direct interactions between the CtBP1 C-terminal end and FANCA, FANCC, FANCF, FANCG, and FANCL were observed by yeast 2-hybrid assays. These interactions were confirmed by co-immunoprecipitation of overexpressed and endogenous full-length proteins in human cells. The fact that CtBP1 binds to components of the FA core complex implies that CtBP1 might function in concert with this protein complex.
A function attributed to the FA core complex is its ubiquitin ligase activity in response to DNA crosslink–induced damage. Mutations or depletion in any of the FA core complex components results in loss of integrity and ubiquitin ligase activity of the core complex.2 Because CtBP1 interacted with several core complex components, we investigated whether it was required for this activity. Experiments with CtBP1-deficient cells revealed that lack of CtBP1 did not impinge on the integrity and ubiquitin ligase activity of the FA core complex; this was demonstrated via co-immunoprecipitation of FA core complex components and robust FANCD2 monoubiquitination and foci formation in response to DNA crosslink damage. However, CtBP1 was found to be essential for cell proliferation, cell survival, and chromosome stability. These results indicate that CtBP1 is not required for events upstream of the FANCD2 signal but may play a role downstream or independent of this FA core complex activity. Previous reports have shown that CtBP1−/− cells are sensitive to a broad range of agents, including hydroxyurea, ultraviolet radiation, and topoisomerase inhibitors.20 These cellular phenotypes are reminiscent of cells deficient in downstream FA proteins such as FANCM-deficient cells.32,33 Cells with mutations in the FANCM gene were also shown to have increased RAD51 foci formation. Consistent with previous studies,34 we observed an increase in RAD51 foci formation in CtBP1i cells after MMC treatment (supplemental Figure 5). These phenotypic similarities between CtBP1-depleted cells and FA-deficient cells imply that CtBP1 could be another FA-related gene acting downstream of the FANCD2-monoubiquitination signal. Hence, we screened seven unassigned lymphoblast or fibroblast lines derived from FA patients. These cells had previously been excluded from belonging to any of the reported subgroups and were subjected to immunoblotting and Sanger sequencing; however, no mutations were identified in CtBP1 or its homolog, CtBP2 (data not shown). Nevertheless, our observations suggest that, like FA proteins, CtBP1 contributes to the maintenance of chromosomal integrity. This function may depend on its interaction with FA proteins and/or may be linked to its primary role in transcriptional regulation. Accordingly, we previously have shown a role of the FA core complex in transcriptional regulation through its interaction with the Notch1 transcriptional repressor, hairy enhancer of split-1 (HES1).24,35 Thus, the FA core complex interaction with CtBP1 may modulate its transcriptional repression activity similar to that observed with HES1. Consequently, depletion of CtBP1 may alter the expression of genes essential for the maintenance of chromosome integrity. A previous report suggested that CtBP1 regulates BRCA1 transcription by directly binding to the BRCA1 promoter in a redox-dependent manner.34 However, in our microarray analysis of CtBPi cells, we did not detect changes in the expression of BRCA1 or other genes involved in crosslink repair. Among the genes up- and downregulated found in CtBP- and FA-depleted cells, genes involved in Wnt activation, such as LRP1, LRP8, DIXDC1, CSNK1G1, FOXM1 and TCF19, were downregulated, whereas Wnt-negative modulators such as DKK1 and TLE4 were upregulated in both cell lines, DKK1 being the most dramatically upregulated gene in both cell lines, specifically in FANCD2-depleted cells. DKK1 upregulation at mRNA and protein levels observed in cells correlate with elevated levels found in sera from FancA−/− and FancC−/− knockout mice.
DKK1 belongs to the Dickkopf family of secreted molecules, which binds or competes with Wnt receptors LRP5/6 and Kremen and prevents Wnt protein signaling,36,37 thus serving as an antagonist of the Wnt signal. DKK1 was initially characterized for its role in developmental morphogenesis of the head, eyes, limbs, and vertebrae; it has been implicated in the onset of polysyndactyly, fused vertebrae, and severe cranial defects associated with anophthalmia in Dkk1−/− mice.38-41 Other studies revealed that DKK1 functions during pituitary and urogenital development.42,43 It is well documented that patients with FA have congenital abnormalities of various organs, including the eyes, kidneys, head, and limbs; absent radii and polysyndactyly are hallmarks of FA.1 In addition, DKK1 has been reported to be overexpressed in many cancers, including multiple myeloma, hepatoblastoma, Wilms’ tumors, and breast, head, neck, and oral cancers.44 These types of cancers often occur in patients with FA (eg, head and neck squamous cell carcinomas are among the most common solid tumors found in FA patients45,46 ), thus making DKK1 a putative player in FA pathogenesis.
In addition, work by Fleming et al47 showed that elevated Dkk1 expression using a transgenic Dkk1 mouse model resulted in increased cell cycling of hematopoietic stem cells (HSCs) and a progressive decline in the regenerative function of HSCs after transplantation. We and others have previously described increased cycling activity and defective reconstitution potential of FA-deficient HSCs.48-51 Consequently, elevated Dkk1 serum levels observed in FancA−/− and FancC−/− knockout mice may be associated with HSC defects found in these FA mice, although this needs to be further investigated.
Because CtBP1 and FA proteins interact and seem to be implicated in the regulation of the Wnt signal molecule DKK1, and because DKK1 is implicated in biological processes linked to FA pathogenesis, we argue that CtBP1 and FA proteins (or pathway) might share a common function. Recent work by Dao et al52 showing that FANCL ubiquitinates the Wnt transcription factor β-catenin supports our finding of interplay between the FA and Wnt pathways. We argue that an altered FA/CtBP1/DKK1 pathway would lead to the reduced capacity for self-renewal exhibited by FA mutant HSCs, which facilitates the progressive loss of bone marrow cells and subsequent bone marrow failure. Further characterization of this pathway and its impact on hematopoiesis will be necessary to understand the pathology of FA and to develop new therapeutic strategies to combat it.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Ms. Chantal Godin for technical expertise. We wish to thank Dr Ezequiel Calvo for analysis of microarray data. We also thank Dr. G. Chinnadurai (St. Louis University Health Sciences Center) for providing the pRc/CMV-T7-CtBP1 construct and Dr. M. Hoatlin (Oregon Health & Science University) for providing anti-FANCC 8F3 monoclonal antibodies.
This work was supported in part by grants from the Canadian Institutes of Health Research (CIHR; MOP-79454) (M.C.), the Canadian Blood Services /CIHR Blood Utilisation and Conservation Initiative grant (M.C. and G.L.), and scholarships from Natural Sciences and Engineering Research of Canada (C.C.H.), Fonds de la Recherche en Santé du Québec (FRSQ) (C.C.H.), CIHR in partnership with the Canadian Fanconi Anemia Research Fund (C.C.H.), the Foundation of Stars (C.C.H. and C.S.T.), and FRSQ senior investigator awards (G.L. and M.C.).
Authorship
Contribution: C.C.H. performed the majority of the experiments, analyzed the data, prepared the figures, and wrote the manuscript. C.S.T. performed co-immunoprecipitation experiments. M.-C.D. performed experiments. K.H. performed screening analyses of cells derived from Fanconi anemia patients. D.S. and G.L. directed some of the research. M.C. directed the research, analyzed the data, and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Madeleine Carreau, Pédiatrie, Centre de Recherche du Centre hospitalier universitaire de Québec, 2705 Boul. Laurier, P-09800, Québec, QC, Canada, G1V 4G2; e-mail: madeleine.carreau@crchul.ulaval.ca