Key Points
UTX regulates migration and hematopoiesis.
Female UTX-KO mice show key features of myelodysplastic syndrome with chromosomal instability.
Abstract
Regulated migration of hematopoietic stem cells is fundamental for hematopoiesis. The molecular mechanisms underlying stem cell trafficking are poorly defined. Based on a short hairpin RNA library and stromal cell-derived factor-1 (SDF-1) migration screening assay, we identified the histone 3 lysine 27 demethylase UTX (Kdm6a) as a novel regulator for hematopoietic cell migration. Using hematopoietic stem and progenitor cells from our conditional UTX knockout (KO) mice, we were able to confirm the regulatory function of UTX on cell migration. Moreover, adult female conditional UTX KO mice displayed myelodysplasia and splenic erythropoiesis, whereas UTX KO males showed no phenotype. During development, all UTX KO female and a portion of UTX KO male embryos developed a cardiac defect, cranioschisis, and died in utero. Therefore, UTY, the male homolog of UTX, can compensate for UTX in adults and partially during development. Additionally, we found that UTX knockdown in zebrafish significantly impairs SDF-1/CXCR4–dependent migration of primordial germ cells. Our data suggest that UTX is a critical regulator for stem cell migration and hematopoiesis.
Introduction
Epigenetic regulators such as histone methyltransferases and demethylases have been identified as critical regulators for normal hematopoiesis and leukemic stem cells.1 Methylation of the histone 3 lysine 27 residue (H3K27) is accomplished by Ezh2 (enhancer of zeste homolog 2), a member of the PRC2 (polycomb repressive complex 2) complex.2 Tri- and di-methylated H3K27 residues (H3K27me3/me2) are associated with transcriptionally repressed chromatin regions.3 Reversion to mono-methylated H3K27 residues (H3K27me1) is facilitated by two proteins: UTX (ubiquitously transcribed tetratricopeptide repeat, X chromosome, Kdm6a) and JMJD3 (Jumonji domain-containing protein 3′, Kdm6b).4-6 Both UTX and JMJD3 contain a JmjC domain, which is required for histone demethylase activity.5 UTX is distinct from JMJD3 by its tetratricopeptide repeat region, which is likely to mediate protein-protein interactions for the assembly of multiprotein complexes. UTY, the male homolog of UTX, was enzymatically inactive when tested in vitro and in vivo.5,7 UTX has been shown to be important for several developmental processes. UTX demethylates H3K27 at the Hox loci,6 regulates lifespan in Caenorhabditis elegans,8 controls posterior development of zebrafish,4 and is a key factor for embryonic development. UTX knockout (KO) null mice have neuronal defects and abnormal cardiac development and are embryonic lethal.9-11
UTX is highly expressed in hematopoietic stem and progenitor cells, with lower relative expression in lineage-dedicated precursors.12 Inactivating mutations in the UTX gene have been identified in human cancers, including multiple myeloma, myeloid leukemia, chronic myelomonocytic leukemia, and glioblastoma.13,14
Using a short hairpin RNA (shRNA) library screening assay, we identified UTX as a new potential regulator of hematopoietic stem cell migration. The impact of UTX on cell migration was confirmed in human Jurkat T-cells, murine 32D cells, and primary human CD34+ hematopoietic stem and progenitor cells (HSPCs). UTX KO leads to growth retardation, cranioschisis, and abnormal cardiac development and is embryonic lethal for homozygous KO females. HSPCs from adult conditional UTX KO mice demonstrated strongly reduced cell migration. Of note, adult female mice with induced UTX KO show myelodysplasia with suppressed erythro-megakaryocytopoiesis and extramedullary compensation in the spleen, highlighting the relevance of UTX for hematopoiesis.
Methods
Production of viral-vector particles containing the shRNA library and transduction of the murine 32D cell line
The murine pRetroSuperCam shRNA library targets approximately 15 000 murine genes.15 pRetroSuperCam vector without shRNA insert was used as control. To generate library containing virus-vector particles, 293T cells were transfected using polyethylenimine. Transduction of 32D cells was performed as described elsewhere.16
Identification of migration-relevant genes using an shRNA library screening assay
The transduction rate of 32D cells was limited to 25% to minimize the number of vector copy inserts per cell. The migration assays were initiated 3 days after transduction. Stromal cell-derived factor-1 (SDF-1) containing RPMI complete medium was added to the lower chamber. Nonmigrated cells were harvested from the upper chamber. After 24 hours of culture, the nonmigrated subgroup of 32D cells was reapplied in congruent migrations assays. After 18 rounds of migration, nonmigrated cells were harvested. A subset of the shRNA library transduced 32D cells was cultured with daily medium changes only. Finally, single cell clones were established by limiting dilution both from enriched nonmigrating and cultured-only library-transduced 32D cells.
Identification of the integrated shRNA to detect potential migration-relevant genes
Integrated shRNA was identified from generated single cell clones. Genomic DNA was isolated using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Single cell clone–specific shRNA-containing polymerase chain reaction (PCR) fragments were amplified and used for nested PCR. Nested PCR products were used for standard sequencing.
Generation of lentiviral vectors containing single shRNA or UTX CDS
pLKO.1 vectors were obtained from OpenBiosystems (Illumina, San Diego, CA). The human UTX coding sequence was cloned into the lentiviral transfer vector pRRL.SIN.cPPT.SFFV.GFP.WPRE (kindly provided by Christopher Baum, Hannover, Germany).17 To produce lentiviral-vector particles, HEK293T cells were transfected with the lentiviral transfer vectors using polyethylenimine.18 Target cells were infected with lentiviral vector particles overnight in the presence of protamine.
Preparation of murine bone marrow, spleen, and peripheral blood
Peripheral blood was taken from anesthetized mice. To collect bone marrow and spleen, mice were euthanized by cervical dislocation. Bone marrow was collected by flushing femurs and tibia. Isolated whole spleen was mashed using a 100-µm nylon mesh to obtain a single cell suspension.
Isolation of human and murine HSPCs
Primary human HSPCs were isolated from cord blood of healthy donors after informed consent in accordance with the Declaration of Helsinki and the approval of the local ethics committee. CD34+ HSPCs were purified using antibody-conjugated magnetic beads (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer’s instructions. Human HSPCs were cultured in X-VIVO10 supplemented with cytokines.
Murine primary HSPCs were isolated from mice using the Lineage Cell Depletion Kit and CD117 MicroBeads (Miltenyi Biotec). Murine HSPCs were cultured in StemSpan SFEM (Stemcell Technologies, Grenoble, France) supplemented with cytokines.
Next-generation sequencing and data analysis
Purified mRNA was used for preparation of a strand-specific RNA sequencing library. Sample libraries were pooled for 75-bp single read sequencing on Illumina HiSequation 2000 (Illumina, San Diego, CA) resulting in 48 to 68 million reads per sample. For data analyses, a splice junction library was created based on known exon-exon junctions according to the Ensembl Genes annotation (v. 67, May 2012).19 Alignment of the reads to the mm9 transcriptome was performed with Parallel Burrows-Wheeler Aligner tool.20 Test for differential gene expression was performed with DESeq. P values for statistical significance of the fold change were adjusted for multiple testing with Benjamini-Hochberg correction for controlling the false discovery rate. Gene expression data were analyzed using the Babelomics suite (V3.2).21
Primordial germ cell migration in zebrafish
UTXLI morpholinos were synthesized by Gene Tools (Philomath, OR). Wobble-mutated capped UTXLI-CDS mRNA was synthesized in vitro. Morpholinos and UTXLI RNA, respectively, were injected into the cytoplasm of 1-cell stage zebrafish embryos. Vasa mRNA in situ hybridization of zebrafish embryos was performed as described previously.22
Colony-forming cell assay
Murine bone marrow was collected from femurs and tibia. Bone marrow cells were resuspended in methylcellulose containing media. After 14 days, hematopoietic colonies were classified and quantified.
Transwell migration assay
Migration assays were carried out using the ChemoTx system (Neuro Probe, Warwickshire, United Kingdom). SDF-1 (R&D Systems, Wiesbaden-Nordenstadt, Germany) was added to the lower chamber. Cells were placed on the top of the membrane. Migrated cells were collected from the bottom chamber and counted using flow cytometry with fixed time settings.
Flow cytometry
To analyze surface expression only, cells were incubated with corresponding antibodies. For comparative analyses of total and surface CXCR4 expression, cells were incubated initially with species-specific CXCR4 antibody in Cytofix/Cytoperm wash buffer (BD Biosciences, Heidelberg, Germany). Subsequently, a subset of cells was permeabilized and fixed with Cytofix/Cytoperm solution to enable analysis of total CXCR4 expression. Again, cells were incubated with species-specific CXCR4 antibody. All samples were analyzed by flow cytometry and evaluated using FlowJo software (Tree Star Inc., Ashland, OR).
To monitor the division history, cells were labeled with carboxyfluorescein succinimidyl ester (CFSE) cell tracker dye (Life Technologies, Darmstadt, Germany) according to manufacturer’s instructions. CFSE-labeled cells were analyzed by flow cytometry.
Quantitative reverse transcriptase polymerase chain reaction
Total RNA was isolated with the RNeasy Mini Kit (Qiagen) and reverse transcribed using Superscript II (Life Technologies) according to the manufacturer’s instructions. Expression levels of genes were quantified by gene-specific TaqMan assays and normalized to gapdh expression.
Chromatin immunoprecipitation
Cells were harvested and washed with ice-cold phosphate-buffered saline. After formaldehyde-based cross-linking, the DNA-protein complex was sheared by ultrasonification. Antibodies (histone 3, H3K27me1, and me3; Millipore, Schwalbach, Germany) for immunoprecipitation were used according to the manufacturer’s instructions. Immunoprecipitated genomic DNA was isolated by phenol/chloroform extraction and ethanol precipitation. CXCR4-specific promoter regions were quantified by quantitative polymerase chain reaction.
Generation of UTX-specific antibody
A novel monoclonal anti-UTX (UTX-DD1) antibody was established by the hybridoma technique as described previously.23 A C-terminal UTX-specific peptide (GenScript, Piscataway, NJ) was covalently linked to keyhole limpet hemocyanin. Initial immunization of UTX KO male mice was followed by 3 boosts every 3 weeks. After isolation, spleen cells were fused with myeloma cells. After hybridoma selection using hypoxanthine-aminopterin-thymidine media, cell culture supernatants of the resulting clones were screened by enzyme-linked immunosorbent assay. Respective hybridomas were subcloned, and single cell clones were established.
Western blot
Tissues and cultured cells were lysed with RIPA buffer. Protein was loaded onto sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels and transferred to nitrocellulose membranes. Membranes were blocked with 5% milk in 1× PBS (+0.05% Tween-20) and incubated with antibodies (histone 3, H3K27me3: Millipore; β-actin: Sigma-Aldrich, Munich, Germany; UTX-DD1) at 4°C overnight, followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibody (anti-mouse IgG-HRP, anti-rabbit IgG-HRP: GE Healthcare, Munich, Germany) before development using Lumi-Light Plus western blot detection system (Roche, Mannheim, Germany).
Generation of UTX-knockout ES cells
The UTX gene was targeted in mouse R1 embryonic stem (ES) cells using a version of the multipurpose allele strategy as described by Testa et al.24 The targeting construct was generated using recombineering technology (Red/ET; supplemental Figure 1A on the Blood website).25 R1 ES cells were cultured on mitomycin C–inactivated mouse embryonic fibroblasts and electroporated with linearized targeting construct using standard conditions, and cells were selected with G418. Colonies were screened for correct targeting events by Southern blot hybridization using an internal and 5′- and 3′-flanking external probes (supplemental Figure 1B). For removing the selection cassettes, correctly targeted clones were coelectroporated with a CAG-Flpo-IRES-puro26 and a CAG-Dre-IRES-puro27 expression vector. Clones were picked 12 days later and screened by polymerase chain reaction for complete recombination and for sensitivity to G418 and hygromycin.
Generation of UTX KO mice and breedings
Correctly targeted clones were injected into C57Bl/6J blastocysts in the MPI-CBG transgenic core facility. Male chimeras were crossed to C57Bl/6J female, and progeny was genotyped for germ line transmission. Female mice carrying the conditional UTX KO construct (UTXFD) were crossed with the ubiquitously expressed Pgk1-Cre deleter (Tg[Pgk1-cre]1Lni) mice.28
For conditional mutagenesis, the UTXFD strain was crossed with the Rosa-Cre-ERT2 strain (C57BL/6-Gt[ROSA]26Sortm9(Cre/ESR1)Arte).29 For induction of UTX KO in adult animals, Tamoxifen was administered orally for 5 consecutive days to ≥11-week-old mice. Experiments were performed with permission from the local authorities.
A complete and detailed description of methods can be found in supplemental Methods.
Approval was received by the ethical committee of the Technische Universitaet Dresden for human HSPCs (permit numbers: EK47022007, EK221102004) and for experimentation with mice (permit number: 24-9168.25-1/2010-7).
Results
Identification of potential migration-relevant genes using an shRNA screening assay
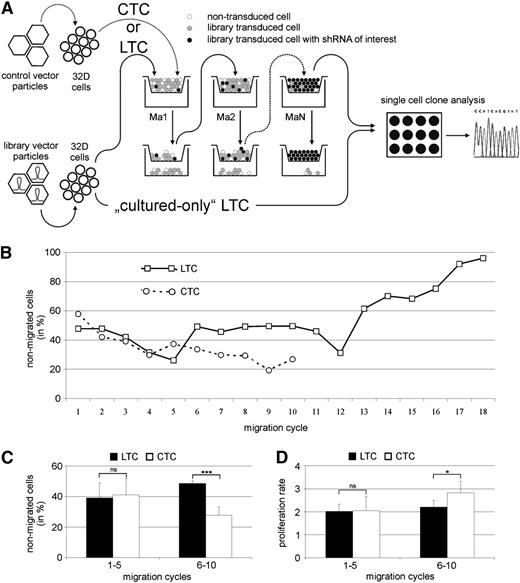
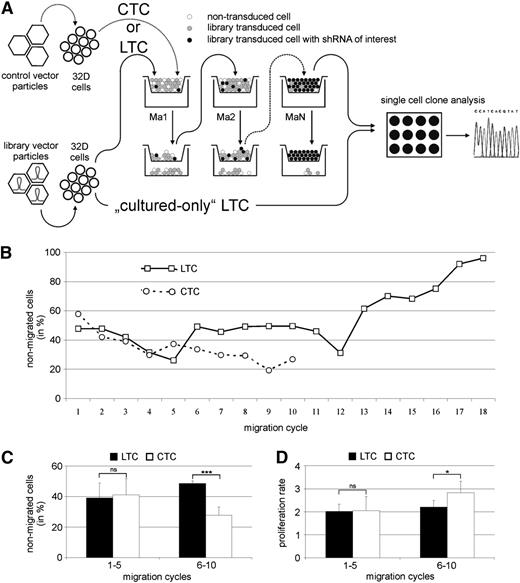
To identify new regulators of SDF-1/CXCR4–dependent cell migration, we performed a transwell migration screening assay to enrich for cells that lost their migration capability because of shRNA expression (Figure 1A).
shRNA library screening assay to identify new regulators of hematopoietic cell migration. (A) The murine 32D cell line was transduced with library containing (LTC) or control (CTC) vector particles. shRNA library transduced and control transduced cells were selected based on the migratory capacity (Ma1). The nonmigrating cells were selected and used for migration on 18 consecutive days (MaN). shRNAs were identified in expanded single cell clones of the nonmigrated LTC by standard sequencing. A cultured-only LTC population was used as control. (B) After 18 rounds of migration, the population of nonmigrating LTC had increased to 96%. CTC demonstrated a constant high migration rate, resulting in too few remaining nonmigrated cells to continue the assay. (C) Although no significant difference in migration was observed between LTC and CTC during the first 5 migration cycles, the percent of nonmigrating cells significantly increased in the LTC during migration cycles 6 to 10. (D) CTC and LTC showed no significant difference in cell proliferation during migration cycles 1 to 5. During the next 5 migration cycles, the proliferation rate of CTC increased compared with LTC. *P < .05, ***P < .001. ns, not significant.
shRNA library screening assay to identify new regulators of hematopoietic cell migration. (A) The murine 32D cell line was transduced with library containing (LTC) or control (CTC) vector particles. shRNA library transduced and control transduced cells were selected based on the migratory capacity (Ma1). The nonmigrating cells were selected and used for migration on 18 consecutive days (MaN). shRNAs were identified in expanded single cell clones of the nonmigrated LTC by standard sequencing. A cultured-only LTC population was used as control. (B) After 18 rounds of migration, the population of nonmigrating LTC had increased to 96%. CTC demonstrated a constant high migration rate, resulting in too few remaining nonmigrated cells to continue the assay. (C) Although no significant difference in migration was observed between LTC and CTC during the first 5 migration cycles, the percent of nonmigrating cells significantly increased in the LTC during migration cycles 6 to 10. (D) CTC and LTC showed no significant difference in cell proliferation during migration cycles 1 to 5. During the next 5 migration cycles, the proliferation rate of CTC increased compared with LTC. *P < .05, ***P < .001. ns, not significant.
We transduced cells of the murine hematopoietic progenitor cell line 32D30 with a retroviral shRNA library encompassing sequences targeting 15 000 murine genes (library transduced cells [LTCs]). One subset of these LTCs was cultured as a proliferation control without migration assays. In addition, we transduced 32D cells with virus-vector particles generated from empty pRetroSuperCam vector (control transduced cells [CTCs]).
Three days after transduction, cells were used for the initial migration assay. During the first 5 migration cycles, no significant difference in migration behavior between LTC and CTC populations was observed (Figure 1B-C). Between migration assay 6 and 10, the proportion of nonmigrating LTCs significantly increased compared with CTCs. Selection of nonmigrating cells for the CTC population had to be stopped after the 10th round of migration because of too few remaining cells resulting from a constantly high migration rate. After migration cycle 12, strongly increasing percentages of nonmigrating LTCs were observed, leading to a final cell population with 96% nonmigrating cells after the 18th round of migration (Figure 1B). LTCs and CTCs displayed no difference in the cell proliferation during migration cycles 1 to 5, whereas during migration cycles 6 to 10, CTCs showed an increased proliferation rate (Figure 1D).
After 18 rounds of migration, we generated single-cell clones from both the nonmigrated and the cultured-only LTC population. From each population, we sequenced polymerase chain reaction products of 120 puromycin-resistant single-cell clones to identify the transduced shRNA. Twenty-four of 120 clones from the nonmigrating LTC population were found to contain shRNA targeting the UTX mRNA sequence, corresponding to a total of 19.8% of identified shRNA sequences. No UTX shRNA-containing cell clone was detected among all single-cell clones of the cultured-only LTC population.
UTX regulates hematopoietic cell migration
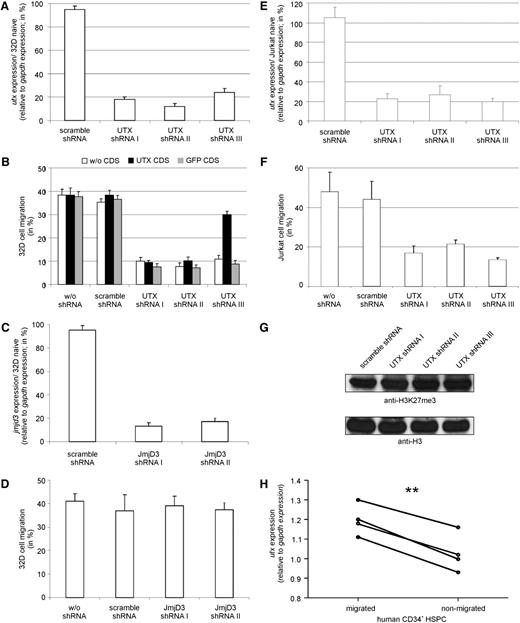
To confirm a requirement for UTX in hematopoietic cell migration, we transduced 32D cells with lentiviral-vector particles containing different shRNAs targeting UTX. All three UTX shRNA-containing cell populations showed a down-regulation of UTX expression compared with naïve and control-transduced 32D cells (Figure 2A). Reduced UTX expression was accompanied by a reduced migration capability (Figure 2B). The shRNA-I and shRNA-II target UTX coding sequence, whereas shRNA-III is specific for the 3′-untranslated region (UTR). Therefore, reduced migration capability could only be rescued by reexpression of a human UTX coding sequence (CDS) lacking the 3′UTR in 32D cells that were transduced with shRNA-III. Migration deficiency could not be rescued by GFP overexpression. Successful rescue using shRNA targeting UTX confirms a specific requirement of UTX for hematopoietic cell migration.
Validation of the regulatory role of UTX on hematopoietic cell migration. (A) UTX shRNA transduced 32D cells showed a significant reduction in UTX expression compared with scramble shRNA transduced 32D cells. (B) 32D cells containing UTX-specific shRNA demonstrated reduced migration compared with wildtype or scramble shRNA transduced 32D cells. Co-overexpression of UTX CDS rescued the migratory phenotype in 32D cells containing shRNA targeting 3′ UTR of UTX mRNA (shRNA-III) but not in 32D cells expressing UTX CDS specific shRNA (shRNA-I, shRNA-II). Coexpression of GFP CDS did not alter migration. (C) JMJD3 shRNA transduced 32D cells showed a significant reduction in JMJD3 expression compared with scramble shRNA transduced 32D cells. (D) 32D cells containing JMJD3 specific shRNA demonstrated no altered migration compared with wildtype or scramble shRNA transduced 32D cells. (E) UTX shRNA transduced Jurkat cells showed a significant reduction in UTX expression compared with scramble shRNA transduced Jurkat cells. (F) Jurkat cells containing UTX-specific shRNA demonstrated reduced migration compared with wildtype or scramble shRNA transduced 32D cells. (G) UTX shRNA transduced Jurkat cells showed no alteration in global level of tri-methylated histone 3 lysine 27 residue (H3K27me3) relative to histone 3 (H3) level compared with scramble shRNA transduced Jurkat cells. (H) Migrated human primary CD34+ hematopoietic stem and progenitor cells (HSPC) of 4 different donors showed significantly higher UTX expression levels than nonmigrated cells. **P < .01
Validation of the regulatory role of UTX on hematopoietic cell migration. (A) UTX shRNA transduced 32D cells showed a significant reduction in UTX expression compared with scramble shRNA transduced 32D cells. (B) 32D cells containing UTX-specific shRNA demonstrated reduced migration compared with wildtype or scramble shRNA transduced 32D cells. Co-overexpression of UTX CDS rescued the migratory phenotype in 32D cells containing shRNA targeting 3′ UTR of UTX mRNA (shRNA-III) but not in 32D cells expressing UTX CDS specific shRNA (shRNA-I, shRNA-II). Coexpression of GFP CDS did not alter migration. (C) JMJD3 shRNA transduced 32D cells showed a significant reduction in JMJD3 expression compared with scramble shRNA transduced 32D cells. (D) 32D cells containing JMJD3 specific shRNA demonstrated no altered migration compared with wildtype or scramble shRNA transduced 32D cells. (E) UTX shRNA transduced Jurkat cells showed a significant reduction in UTX expression compared with scramble shRNA transduced Jurkat cells. (F) Jurkat cells containing UTX-specific shRNA demonstrated reduced migration compared with wildtype or scramble shRNA transduced 32D cells. (G) UTX shRNA transduced Jurkat cells showed no alteration in global level of tri-methylated histone 3 lysine 27 residue (H3K27me3) relative to histone 3 (H3) level compared with scramble shRNA transduced Jurkat cells. (H) Migrated human primary CD34+ hematopoietic stem and progenitor cells (HSPC) of 4 different donors showed significantly higher UTX expression levels than nonmigrated cells. **P < .01
Notably, 32D cells expressing JMJD3 shRNAs showed efficient reduction of JMJD3 mRNA levels (Figure 2C) but no reduction of migration potential compared with control-transduced and naïve 32D cells (Figure 2D).
Expression of UTX shRNAs in cells of the T-cell leukemic Jurkat31 cell line resulted in reduced UTX expression level and a reduced migration potential compared with control-transduced cells (Figure 2E-F). The global level of tri-methylated histone 3 lysine 27 residue (H3K27me3) showed no alteration after UTX knockdown (Figure 2G).
Proliferation of UTX knockdown Jurkat cells as measured by CFSE staining and cell counting was not altered compared with control cells (supplemental Figure 2).
To analyze UTX expression in human HSPCs, we separated HSPCs from 4 different donors according to their migration potential using an SDF-1–dependent migration assay. Consistent with our cell line data, migrated human primary HSPCs showed a significantly higher UTX expression level compared with nonmigrated cells (Figure 2H).
CXCR4 expression and CXCR4 localization are not influenced by UTX
We analyzed CXCR4 expression, CXCR4 localization, and H3K27 methylation of the CXCR4 promoter to test whether the reduced migration of UTX shRNA expressing cells is caused by an altered expression or localization of the SDF-1–specific receptor CXCR4 (supplemental Figure 3A).
UTX knockdown did not influence CXCR4 expression or localization in both 32D and Jurkat cells as measured by quantitative reverse transcriptase-polymerase chain reaction or flow cytometry (supplemental Figure 3B). Furthermore, quantification of the H3K27 methylation status of the CXCR4 promoter region of UTX shRNA-expressing 32D cells demonstrated equal H3K27me1 and H3K27me3 methylation in 32D cells expressing UTX or scramble shRNAs (supplemental Figure 3C).
Primordial germ cell migration in zebrafish
To test the relevance of UTX on SDF-1/CXCR4–dependent cell migration in vivo, we studied migration of primordial germ cells (PGCs) during embryonic development of zebrafish (Figure 3). PGCs follow a complex path of migration throughout the early embryo within the first 24 hours of development, which is primarily controlled by dynamic changes in the SDF-1 expression pattern.32
UTX expression is critical for PGC migration in zebrafish. (A) Treatment of zebrafish embryos at the 1-cell stage with UTX-specific morpholinos (UTX MO) resulted in significantly reduced migration of PGCs to the main cluster. Coinjection of MO-resistant UTX mRNA significantly restored migration of PGC to the main cluster. (B) Representative images of zebrafish embryos injected at the 1-cell stage with control MO, UTX MO, or UTX MO + MO-resistant UTX mRNA and stained for Vasa mRNA 24 hours after fertilization in situ. Red arrowhead indicates the normal position of the germ cell cluster in the embryo, whereas black arrowheads show ectopic positions. ***P < .001. MO, morpholino; ns, not significant.
UTX expression is critical for PGC migration in zebrafish. (A) Treatment of zebrafish embryos at the 1-cell stage with UTX-specific morpholinos (UTX MO) resulted in significantly reduced migration of PGCs to the main cluster. Coinjection of MO-resistant UTX mRNA significantly restored migration of PGC to the main cluster. (B) Representative images of zebrafish embryos injected at the 1-cell stage with control MO, UTX MO, or UTX MO + MO-resistant UTX mRNA and stained for Vasa mRNA 24 hours after fertilization in situ. Red arrowhead indicates the normal position of the germ cell cluster in the embryo, whereas black arrowheads show ectopic positions. ***P < .001. MO, morpholino; ns, not significant.
Injection of either one of two antisense morpholino oligonucleotides (MOs) specific against UTX into 1-cell stage zebrafish embryos resulted in a significantly reduced migration of PGCs to their bilateral location at the position of the future gonads at 24 hours after fertilization (main cluster) compared with zebrafish embryos injected with control MOs. The number of PGCs remaining at ectopic regions was strikingly reduced when UTX mRNA that could not be bound by the MOs was coinjected, restoring the natural migration to the main cluster. Confirming previously described essential developmental functions of UTX,4 MO-injected embryos died at later stages of development, whereas coinjected (rescued) embryos survived at a comparable rate as control-injected embryos (data not shown).
UTX KO female mice die before birth
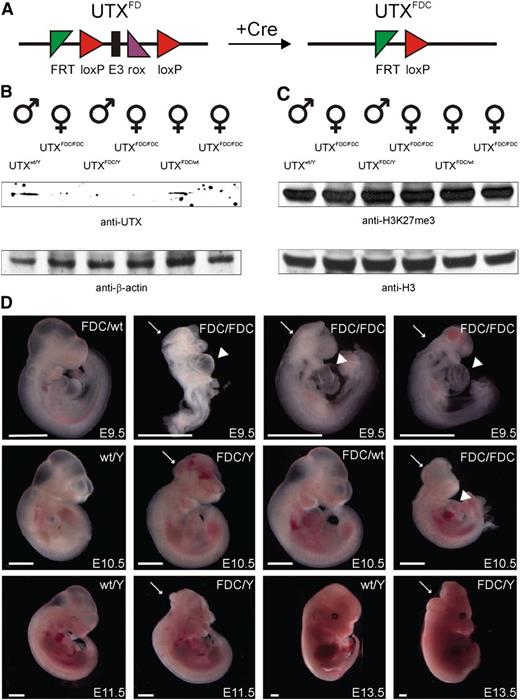
To further investigate the function of UTX in vivo, we generated murine KO ES cells and mice using the multipurpose allele strategy.24 A transcriptional stop cassette flanked by flippase recognition target sites was inserted into intron 2 upstream of the frameshifting exon 3 of UTX, which was flanked by loxP sites and followed by an hygromycin selection cassette flanked by rox sites in intron 3 (supplemental Figure 1). Because UTX is located on the X chromosome and the R1 ES cells are male, the transcriptional stop cassette inserted into intron 2 leads to complete UTX KO (knockout-first). We did not obtain chimeric mice after injection of a clone that carried the knockout-first allele. To overcome any potential viability problems of the UTX KO cells after injection into blastocysts, we removed the selection cassettes using Flpo- and Dre-mediated recombination in vitro.26,27 This genetic manipulation converted the knockout-first allele into conditional, which we termed UTXFD (F, Flp recombination; D, Dre recombination), and led to chimeric mice that contributed to the germ line after crossing with C57Bl/6J females. As expected for an X-linked gene, only female progeny carried the conditional allele.
To induce the frameshifting mutation in vivo, UTXFD/wt female mice were crossed to ubiquitously Cre-expressing Pgk1-Cre-deleter mice.28 Cre-recombination removes the frameshifting exon 3, which results in the UTXFDC (C, Cre recombination) allele with a premature stop codon in exon 4 and subsequent activation of nonsense-mediated decay for mRNA degradation. To our surprise, we obtained UTXFDC/Y male mice that survived to adulthood and were fertile. After intercrossing UTXFDC/Y males with UTXFDC/wt females, we could not detect any UTXFDC/FDC female progeny, indicating that the female homozygous mice die before birth. We also observed a reduced ratio (∼10% instead of 25%) of the UTXFDC/Y males (Table 1). Recently, this deviation from the Mendelian ratio for UTXFDC/Y males was also reported by others.10,11 Interestingly, a subsequent cross of UTXFDC/wt females with C57Bl/6J males produced only one UTXFDC/Y living male (0.8%) out of 119, indicating a variable contribution from the genetic background (supplemental Table 2).
Molecular and morphological phenotype of the UTX KO embryos
To analyze the embryonic phenotype of UTX KO in more detail, we isolated embryos at stages E8.5, E9.5, E10.5, E11.5, and E13.5 from UTXFDC/Y × UTXFDC/wt intercrosses (Table 1; Figure 4A).
UTX expression is essential for embryonic development. (A) Schematic representation of the UTX conditional allele (UTXFD) and the conversion to the frameshifted KO allele (UTXFDC) after excision of exon 3 with Cre recombinase. (B) Western blot of littermate lysates from one UTXFDC/Y × UTXFDC/wt intercross showed efficient knockout of UTX. (C) UTX KO had no influence on the global level of the tri-methylated histone 3 lysine 27 residue (H3K27me3) relative to the histone 3 (H3) level in embryos derived from an UTXFDC/Y × UTXFDC/wt intercross. (D) Embryos isolated at different stages from UTXFDC/Y × UTXFDC/wt intercrosses. At E9.5 and E10.5, female UTXFDC/FDC embryos show growth retardation, cardiac malformations (arrowhead), and neural tube closure defects (arrow) and die between E11.5 and E13.5. A fraction of UTXFDC/Y male embryos exhibit cranioschisis at all analyzed stages. Scale bar, 1 mm.
UTX expression is essential for embryonic development. (A) Schematic representation of the UTX conditional allele (UTXFD) and the conversion to the frameshifted KO allele (UTXFDC) after excision of exon 3 with Cre recombinase. (B) Western blot of littermate lysates from one UTXFDC/Y × UTXFDC/wt intercross showed efficient knockout of UTX. (C) UTX KO had no influence on the global level of the tri-methylated histone 3 lysine 27 residue (H3K27me3) relative to the histone 3 (H3) level in embryos derived from an UTXFDC/Y × UTXFDC/wt intercross. (D) Embryos isolated at different stages from UTXFDC/Y × UTXFDC/wt intercrosses. At E9.5 and E10.5, female UTXFDC/FDC embryos show growth retardation, cardiac malformations (arrowhead), and neural tube closure defects (arrow) and die between E11.5 and E13.5. A fraction of UTXFDC/Y male embryos exhibit cranioschisis at all analyzed stages. Scale bar, 1 mm.
Western blot analyses of single littermates from these crossings revealed a loss of UTX protein in homozygous UTXFDC/FDC female and hemizygous UTXFDC/Y male embryos compared with heterozygous UTXFDC/wt female and UTXwt/Y male embryos (Figure 4B). Interestingly, global H3K27me3 levels in homozygous UTXFDC/FDC female and hemizygous UTXFDC/Y male embryos were similar to global H3K27me3 level in heterozygous UTXFDC/wt female and UTXwt/Y male embryos (Figure 4C).
A portion of UTXFDC/FDC female embryos (∼33%) had an abnormal morphology already at E8.5. At E9.5, all homozygous UTXFDC/FDC females showed pronounced growth retardation, neural tube closure defects (cranioschisis), and cardiac malformations (Figure 4D). Also, some of the UTXFDC/Y male embryos showed the same morphological abnormalities, whereas heterozygous UTXFDC/wt females were not affected macroscopically.
A more severe phenotype was observed at day E10.5. All homozygous UTXFDC/FDC female embryos showed cranioschisis and general growth retardation (Figure 4D). Again, some of the UTXFDC/Y male embryos were morphologically not affected, whereas others showed cranioschisis without growth retardation (∼17%). UTXFDC/FDC female embryos died between E11.5 and E13.5, whereas some of the UTXFDC/Y male embryos exhibited cranioschisis at E13.5 (18%; Figure 4D). Cardiac malformations and neural tube closure defects in UTX KO embryos have been also described by 2 independent groups,2,10 and neural tube closure defects were also observed by a gene-trapped UTX allele,9 thus verifying the phenotype of our UTX KO allele.
Hematopoietic phenotype of adult conditional UTX KO mice
To be able to analyze the role of UTX in adult mice, we crossed the conditional UTXFD mice to the Rosa-Cre-ERT2 line.29 The Cre-ERT2 fusion protein is expressed from the Rosa26 locus, but Cre-ERT2 associates with the hsp90 in the cytoplasm and remains inactive. To induce Cre expression, mice were treated at 11 to 14 weeks of age with Tamoxifen for 5 consecutive days per gavage. Ten days after Tamoxifen treatment, recombination efficiency was tested in different tissues. A complete recombination was observed in all tissues but brain. Brain tissue showed incomplete recombination efficiency because of an insufficient expression of CreERT2 in postmitotic neurons (supplemental Figure 4). If not mentioned otherwise, animals were euthanized 5 weeks after Tamoxifen treatment.
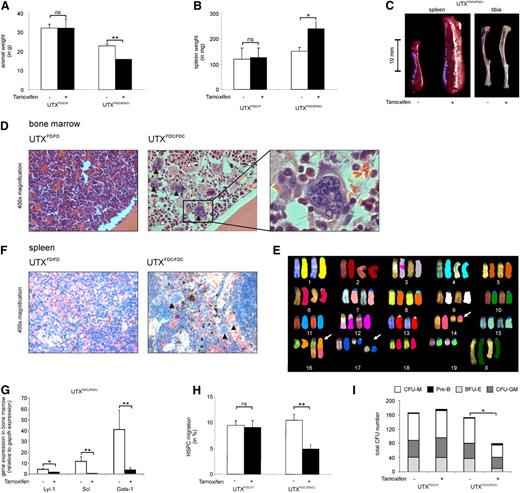
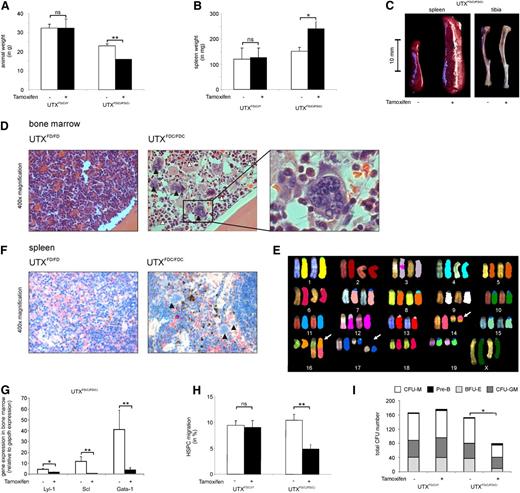
Tamoxifen-treated homozygous UTXFDC/FDC females showed a significant weight loss compared to hemizygous UTXFDC/Y males (Figure 5A). Of note was the strikingly increased spleen weight (Figure 5B-C) and the obvious anemic appearance of the bones (Figure 5C) found in the UTXFDC/FDC female mice compared to the UTXFDC/Y male mice.
UTX expression is critical for HSPC cell migration and normal hematopoiesis. (A) Tamoxifen treatment of adult UTXFDC/FDC female mice results in significant weight loss compared with nontreated animals. Tamoxifen treatment had no effect on the body weight of UTXFDC/Y male mice. (B) Treatment of adult UTXFDC/FDC female mice results in a significant increase of spleen weight compared with nontreated animals. Tamoxifen treatment had no effect on spleen weight of UTXFDC/Y male mice. (C) Treatment of adult UTXFDC/FDC female mice results in anemic bones and a significant increase of spleen size compared with nontreated animals. Representative images of spleen and tibia are shown. (D) Bone marrow sections from adult UTXFDC/FDC female mice showed disturbed cell distribution with concomitant pronounced myeloid dysplasia. Images display an increased number of megakaryocytes with abnormal nuclear lobation (black arrowhead) and granulocytes with nuclear hyposegmentation (white arrowhead) in comparison with sections from UTXFD/FD female mice. Representative images from hematoxylin and eosin staining are shown. (E) Example of metaphase spreads from SKY analysis of UTXFDC/FDC bone marrow cells is given. The following chromosomal aberrations were evident: T(14B;16B?) and double-strand break of chromosome 17 (white arrows). (F) Spleens from adult UTXFDC/FDC female mice showed increased extramedullary hematopoiesis and megakaryocytes with abnormal nuclear lobation (black arrowhead). Representative images from chloracetate-esterase staining are shown. (G) Significantly reduced gene expression levels of the erythroid and megakaryocytic lineage-specific transcription factors LYL-1, SCL, and GATA-1 were observed in the bone marrow of UTXFDC/FDC mice compared with UTXFD/FD female mice. (H) HSPCs from Tamoxifen-treated adult UTXFDC/FDC female mice demonstrated a significantly reduced migration potential compared with HSPCs from nontreated animals. Tamoxifen treatment of UTXFDC/Y male mice did not influence the migration rate of HSPCs. (I) Bone marrow cells from Tamoxifen-treated adult UTXFDC/FDC female mice demonstrated a significantly reduced number of CFUs and an altered composition of CFUs compared to cells from UTXFD/FD, UTXFD/Y, and UTXFDC/Y animals, respectively. *P < .05, **P < .01. BFU-E, burst forming unit–erythroid; CFU-GM, granulocyte-macrophage progenitor; CFU-M, macrophage; ns, not significant; Pre-B, lymphoid progenitors.
UTX expression is critical for HSPC cell migration and normal hematopoiesis. (A) Tamoxifen treatment of adult UTXFDC/FDC female mice results in significant weight loss compared with nontreated animals. Tamoxifen treatment had no effect on the body weight of UTXFDC/Y male mice. (B) Treatment of adult UTXFDC/FDC female mice results in a significant increase of spleen weight compared with nontreated animals. Tamoxifen treatment had no effect on spleen weight of UTXFDC/Y male mice. (C) Treatment of adult UTXFDC/FDC female mice results in anemic bones and a significant increase of spleen size compared with nontreated animals. Representative images of spleen and tibia are shown. (D) Bone marrow sections from adult UTXFDC/FDC female mice showed disturbed cell distribution with concomitant pronounced myeloid dysplasia. Images display an increased number of megakaryocytes with abnormal nuclear lobation (black arrowhead) and granulocytes with nuclear hyposegmentation (white arrowhead) in comparison with sections from UTXFD/FD female mice. Representative images from hematoxylin and eosin staining are shown. (E) Example of metaphase spreads from SKY analysis of UTXFDC/FDC bone marrow cells is given. The following chromosomal aberrations were evident: T(14B;16B?) and double-strand break of chromosome 17 (white arrows). (F) Spleens from adult UTXFDC/FDC female mice showed increased extramedullary hematopoiesis and megakaryocytes with abnormal nuclear lobation (black arrowhead). Representative images from chloracetate-esterase staining are shown. (G) Significantly reduced gene expression levels of the erythroid and megakaryocytic lineage-specific transcription factors LYL-1, SCL, and GATA-1 were observed in the bone marrow of UTXFDC/FDC mice compared with UTXFD/FD female mice. (H) HSPCs from Tamoxifen-treated adult UTXFDC/FDC female mice demonstrated a significantly reduced migration potential compared with HSPCs from nontreated animals. Tamoxifen treatment of UTXFDC/Y male mice did not influence the migration rate of HSPCs. (I) Bone marrow cells from Tamoxifen-treated adult UTXFDC/FDC female mice demonstrated a significantly reduced number of CFUs and an altered composition of CFUs compared to cells from UTXFD/FD, UTXFD/Y, and UTXFDC/Y animals, respectively. *P < .05, **P < .01. BFU-E, burst forming unit–erythroid; CFU-GM, granulocyte-macrophage progenitor; CFU-M, macrophage; ns, not significant; Pre-B, lymphoid progenitors.
Peripheral blood analysis of UTXFDC/FDC females revealed decreased red blood cell counts and significantly reduced hemoglobin levels, as well as significantly reduced mean corpuscular volume of erythrocytes compared with the blood counts of the same animals before Tamoxifen treatment (Table 2). In addition, an absolute thrombocytopenia and a relative leukocytopenia were evident. In contrast, Tamoxifen-treated UTXFDC/Y male mice did not show noticeable differences in blood counts compared to pretreatment.
Analysis of the cell composition of the bone marrow of the UTXFDC/FDC female mice demonstrated a significantly diminished percentage of erythroid cells (CD45−Ter119+; Table 2), in particular a dramatic loss of Ter119+CD71+ precursors accounting for the anemic appearance of the bones (Figure 5C).
Histological analyses of bone marrow sections from UTXFDC/FDC female mice taken 14 days after Tamoxifen treatment showed disturbed cell distribution with pronounced dysplasia and disrupted maturation in the erythroid, megakaryocytic, and granulocytic lineage. In detail, an increased number of megakaryocytes with abnormal nuclear lobation and atypical mitosis, as well as dysplastic granulocytes with nuclear hyposegmentation, were observed (Figure 5D).
Furthermore, blood tests of creatinine, albumin, lactate dehydrogenase, conjugated and total bilirubin, amylase, lipase, alanine-aminotransferase, aspartate-aminotransferase, and haptoglobine in UTXFDC/FDC female mice did not show any differences compared with UTXFD/FD, UTXFDC/Y, and UTXFD/Y mice.
Two weeks after Tamoxifen treatment, the bone marrow of the UTXFDC/FDC females revealed similar total numbers of HSPCs compared with UTXFDC/Y control mice (data not shown). Analyses of differentiation capacity of HSPCs from UTXFDC/FDC females showed a significantly reduced formation of CFUs with concomitant reduction in granulocyte-macrophage progenitor and burst forming unit–erythroid, as well as macrophage colony number, compared with HSPCs from Tamoxifen-treated control mice (Figure 5I).
To test bone marrow cells for chromosomal instability, we performed cytogenetic analyses using spectral karyotyping of two UTXFDC/FDC mice 2 weeks after Tamoxifen treatment. We found chromosomal double-strand breaks and numerical and structural chromosomal aberrations in 9 of 15 and 5 of 15 metaphases, respectively (Figure 5E). Structural chromosomal aberrations included translocations of chromosome fragments, partial loss of chromosomal material, and the formation of dicentric chromosomes. We did not observe recurrent chromosomal aberrations.
The spleen of the UTXFDC/FDC female mice were found to be strikingly enlarged compared with Tamoxifen-treated UTXFDC/Y male mice (Figure 5C), with a strongly increased percentage of erythrocyte precursors indicative of splenic erythropoiesis (Table 2). In addition, histological analyses of the spleen of UTXFDC/FDC female mice showed an increased extramedullary hematopoiesis and numerous megakaryocytes with abnormal nuclear lobation (Figure 5F). Concomitantly, a significant B-lymphocytopenia was found in the spleen (Table 2).
To study the functional mechanism of the observed dysplasia in the erythroid and megakaryocytic lineage, we analyzed the gene expression of the transcription factors GATA-1, LYL-1, and SCL by quantitative reverse transcriptase-polymerase chain reaction in the bone marrow and detected a significant down-regulation of all 3 transcription factors in UTXFDC/FDC mice (Figure 5G). Global gene expression analysis demonstrated significant down-regulation of GATA-1 (UTXFDC/FDC/UTXFD/FD = 0.19; P < .001) and SCL (UTXFDC/FDC/UTXFD/FD = 0.25; P < .001). In addition, KLF1 (UTXFDC/FDC/UTXFD/FD = 0.024; P < .001) and IKAROS (UTXFDC/FDC/UTXFD/FD = 0.56; P < .005) were significantly down-regulated, whereas PU.1 (UTXFDC/FDC/UTXFD/FD = 10.27; P < .001) was significantly up-regulated.
Influence of UTX KO on migration of primary HSPCs
To analyze the influence of UTX on HSPC migration, we isolated HSPCs and found significantly reduced migration of UTXFDC/FDC HSPCs but not of HSPCs from UTXFD/FD, UTXFDC/Y, and UTXFD/Y mice toward SDF-1 (Figure 5H).
To study the functional effect of UTX KO on HSPC migration in more detail, we performed comparative gene expression analyses using next-generation sequencing. Therefore, we used the nonmigrating cell populations of an SDF-1–dependent migration assay with HSPCs from UTXFDC/FDC and UTXFD/FD mice. We identified 3202 genes that displayed significantly different transcript levels between these 2 groups. Using gene ontology classification, we identified 31 genes that are associated with chemotaxis (GO0006935; GO0030334). Furthermore, we identified differentially regulated genes that play a role in microtubule-based processes (GO0007017) and in the alteration of the actin skeleton (GO0030029; supplemental Table 4).
Discussion
Targeted cell migration is essential for many physiological and pathological processes. The molecular mechanisms underlying stem cell locomotion are poorly defined. We performed a genome-scale retroviral shRNA screening assay to discover novel genes implicated in hematopoietic stem cell migration (Figure 1). Among a list of candidate genes, the most frequently detected gene was the H3K27 histone demethylase UTX. UTX has extensively been studied during the past years and is now implicated in organ development.4-6 However, a role for UTX in stem cell migration has not yet been identified. To validate the screen, we confirmed that shRNA-mediated down-regulation of UTX reduced cell migration, whereas reexpression of UTX rescued the migratory phenotype of hematopoietic cell lines (Figure 2). Migration of human HSPCs correlated with endogenous UTX levels because migrated primary HSPCs expressed UTX at higher levels than nonmigrated cells. Next, we found that SDF-1/CXCR4–dependent PGC migration in zebrafish is UTX dependent. Additionally, primary HSPCs from adult conditional UTXFDC/FDC female mice demonstrated significantly reduced migration potential toward SDF-1 compared with HSPCs from UTXFDC/Y male mice. Our observations might suggest a relationship between UTX regulation and SDF-1/CXCR4 stem cell migration. However, we did not find any UTX-mediated regulation of CXCR4 on the level of gene expression, protein distribution, or epigenetic regulation of the CXCR4 promoter (supplemental Figure 3). Gene expression analyses of primary nonmigrated murine UTX KO and wildtype HSPCs using next-generation sequencing revealed a number of cell migration–related genes that are significantly influenced by UTX (supplemental Table 4). Although these analyses provide insight, it remains to be established how UTX regulates migration precisely.
During embryonic development, most UTXFDC/FDC female and all UTXFDC/Y male embryos are normal at E8.5. However, all UTXFDC/FDC female embryos show growth retardation, cardiac malformations, and neural tube closure defects at E9.5. All UTXFDC/FDC embryos die between E11.5 and E13.5, whereas some of the UTXFDC/Y male embryos exhibit cranioschisis at E13.5 (Figure 4; Table 1).
These observations agree with recent publications from 2 independent groups. Welstead et al10 and Lee et al11 reported similar embryonic phenotypes of their UTX KO mice, including cranioschisis and abnormal cardiac development. The neurological defect had been first described by Cox et al,9 who analyzed gene-trapped X-chromosomal genes. Both groups that generated targeted UTX KO obtained either the ES cells11 or the targeting construct10 from the resource of the European Conditional Mouse Mutagenesis Program (EUCOMM).33 Because the EUCOMM alleles are based on our knockout-first design,24 both the EUCOMM UTX-allele and the one used here are very similar, although not identical. Both used a similar conditional design and flanked the critical exon 3 with loxP sites. Despite the use of different Cre-deleters for inducing the frameshift mutation (Ella-Cre,11 Nestin-Cre,10 and Pgk1-Cre in our study), the embryonic phenotype is similar. Remarkably, although hemizygous UTXFDC/Y male embryos have no growth retardation, some also show cranioschisis and occasional abnormal cardiac development and do not survive. Some UTXFDC/Y males survive to adulthood and are fertile. Notably, after an additional backcross of UTXFDC/wt females to C57Bl/6J males, we could obtain only one living UTXFDC/Y male, indicating a contribution from the genetic background (supplemental Table 3).
Because UTX KO females die in utero, we generated an inducible UTX KO mouse and report the adult phenotype of UTX KO mice. Tamoxifen-induced UTX KO resulted in a significant weight loss in UTXFDC/FDC females (Figure 5). Induction of homozygous UTX KO in female mice resulted in bones with an obvious anemic appearance. Red blood cell counts in the peripheral blood were significantly diminished, and erythrocytes and their precursors were found to be nearly absent in the bone marrow (Table 2). Besides the erythrocytopenia, UTXFDC/FDC mice displayed profound thrombocytopenia and relative leukocytopenia. Furthermore, spleen of UTXFDC/FDC mice was significantly enlarged and demonstrated extramedullary erythropoiesis, likely to be a compensatory reaction for the erythrocytopenia seen in bone marrow and peripheral blood. In addition, histologic analyses of the spleen of UTXFDC/FDC mice demonstrated dysplasia in the erythroid and megakaryocytic lineage. Bone marrow sections from these mice showed marked dysplasia in the erythroid, megakaryocytic, and granulocytic lineages. Based on the observed erythrocytopenia and thrombocytopenia, we studied the gene expression of transcriptional regulators, known to be critical for erythropoiesis and megakaryopoiesis,34 and found significant down-regulation of GATA-1, LYL-1, SCL, KLF1, and IKAROS, as well as significant up-regulation of PU.1.34-37 Moreover, analyses of bone marrow cells of the UTX KO mice demonstrated chromosomal instability with marked chromosomal rearrangements. The observed pancytopenia together with the histological findings of myeloid dysplasia in UTXFDC/FDC mice coincide with key features of myelodysplastic syndrome, also seen in GATA-1 knockdown mice.38-40
There is growing evidence supporting the hypothesis that JmjC domain-containing proteins like UTX may serve other purposes in addition to being H3K27 demethylases. Whether UTX-mediated regulation of cell migration requires H3K27 demethylase activity remains to be determined. We observed that knockdown of the other known H3K27 demethylase, JMJD3, did not affect migration in our assay, indicating specificity for UTX and not H3K27 demethylation in general. UTX-1, the unique homolog in C. elegans, is an H3K27me2/me3 demethylase; however, its essential role in embryonic development is demethylase independent.41 Miller et al42 also reported on a H3K27 demethylase-independent function of the JmjC-domain containing proteins in Th1 cells. We analyzed the global level of tri-methylated histone 3 lysine 27 residue (H3K27me3) of embryonic tissue and did not find any change in UTX KO tissue compared with wildtype. Similar findings are reported by Shpargel et al,7 who demonstrated that UTX KO in murine embryonic fibroblasts did not influence H3K27me3 levels, and by Wang et al,43 who demonstrated histone demethylase–independent regulation of mesoderm differentiation by UTX.
We report that induction of UTX-KO has no obvious influence on adult male mice, indicating that UTY, the male counterpart of UTX, provides at least some of the UTX functions (Figure 4 and 5). Moreover, Shpargel et al7 reported that UTY KO mice are viable, whereas UTX/UTY double KO mice are embryonic lethal and phenocopy UTX-KO female mice. Although UTY contains the critical amino acids for catalysis, UTY does not demethylate H3K27, as has been reported by different laboratories.5-7,41,44 Overall, these data suggest that both UTX and UTY can regulate gene activity through histone demethylase–independent mechanisms.
In conclusion, we demonstrated the regulatory role of UTX for stem cell migration. Moreover, we showed that UTX is critical for embryonic development and for hematopoiesis in adult female mice.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank René Bernards and Roderick Beijersbergen for providing the murine shRNA library (Nederlands Kanker Instituut-Antoni van Leeuwenhoek Ziekenhuis, Netherlands); Martin Ryser for guidance at project initiation; and Alexander Platz (Cord Blood Bank of the Deutsche Knochenmarkspenderdatei, University Clinic Dresden, Dresden, Germany) for providing the cord blood samples. Furthermore, the authors thank Mandy Obst, Beate Gnauck, Katrin Navratiel, and Jenny Marzahn for technical assistance, as well as Gustavo B. Baretton for financial support in processing the tissue samples.
This work was supported by the Collaborative Research grant SFB-655 (German Research Foundation [DFG]) (A.D., K.A., F.B., S.B., G.W.), the European Union 7th Framework integrated project SyBoSS (A.F.S.), and DFG grant SPP1356 (K.A.). This study was also supported by a grant from the Wilhelm-Sander-Stiftung (2010.044.1) (M.H.M.).
Authorship
Contribution: S.T., K.A., and S.B. designed and performed the experiments and analyzed the data. S.T., A.F.S., K.A., and S.B. wrote the manuscript; T.G. and C.R. contributed significantly to the laboratory work; and J.B. contributed to the laboratory work. G.Ö. and G.W. performed PGC migration analyses; D.A. and A.D. performed next-generation sequencing analysis; C.J. and M.H.M. performed histological analysis; and J.F., K.A., and A.F.S. generated the UTX knockout ES cells and mice. F.B. was involved in the shRNA screening assay. I.M. and M.B. generated the UTX-specific antibody. B.K. and E.S. performed the cytogenetic analyses. All authors critically reviewed the manuscript and agreed to its publication.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sebastian Brenner, Department of Pediatrics, University Clinic “Carl Gustav Carus”, Fetscherstrasse 74, 01307 Dresden, Germany; e-mail sebastian.brenner@uniklinikum-dresden.de. Konstantinos Anastassiadis, BIOTEC, Technische Universitaet Dresden, Tatzberg 47/49, 01307 Dresden, Germany; e-mail konstantinos.anastassiadis@biotec.tu-dresden.de.
References
Author notes
T.G. and C.R. contributed equally to this study.