Key Points
Newly identified mutations in HIF2A result in polycythemia and neuroendocrine tumors.
Disruption of the hydroxylation domain in HIF-2α results in protein stabilization, pseudohypoxia, and tumorigenesis.
Abstract
Hypoxia-inducible factors (HIFs) control the cellular response to hypoxia and, when dysregulated, contribute to tumorigenesis. Previously, we identified 2 gain-of-function somatic mutations in patients presenting with multiple paragangliomas or somatostatinomas, and polycythemia. Here, we report 2 additional unique HIF2A mutations, which disrupt the hydroxylation domain recognized by prolyl hydroxylase domain-2 containing protein, leading to stabilization of HIF-2α and increased expression of hypoxia-related genes.
Introduction
Hypoxia-inducible factors (HIFs) are central to the cellular hypoxic response.1 HIFs are αβ-heterodimers composed of a constitutively expressed HIF-β subunit and an oxygen-dependent HIF-α subunit. HIF-α is inducible during hypoxic conditions via reduced oxygen-dependent prolyl hydroxylation, decreased binding to the von Hippel-Lindau protein (VHL), and thus decreased proteasomal degradation.2,3 Stabilized HIF-α translocates into the nucleus and leads to an increase in the expression of pro-survival genes such as EPO, EDN1, GLUT1, and VEGFA.4 HIF-α has been found to be critical for vasculogenesis and hematopoiesis during development.5 Abnormal HIF-α function enhances cell proliferation and growth, which is a recognized mechanism in aggressive tumors.6
Polycythemia is a hematologic disorder defined by an increase in red cell mass. Genetic changes in key elements of hypoxia-sensing pathways, such as germline mutations in VHL or EGLN1, are essential for the establishment of polycythemia.7,8 Percy et al9 reported novel HIF2A mutations (M535V, G537R) associated with polycythemia as a result of constitutively activated hypoxic cell signaling. Paragangliomas and somatostatinomas are catecholamine- and somatostatin-producing neuroendocrine tumors, respectively. Mutations in SDHx, SDHAF2, TMEM127, MAX, EGLN1, and VHL have been implicated in the pathogenesis of these neuroendocrine tumors.10 These mutations indirectly lead to HIF stabilization and aberrant expression of hypoxia-related genes. These diseases share a disruption in HIF-α regulation.
Study design
This study was approved by the institutional review board of the Eunice Kennedy Shriver National Institute of Child Health and Human Development. The patients provided written informed consent in accordance with the Declaration of Helsinki.
Peptides, plasmids, and mutagenesis
Wild-type HIF-2α ELDLETLAPYIPMDGEDFQ and HIF-2α mutants (L529P) ELDLETPAPYIPMDGEDFQ and (Y532C) ELDLETLAPCIPMDGEDFQ were synthesized, and the N terminus was biotinylated (Peptide 2.0). HA (hemagglutinin)-HIF-2α-pcDNA3 plasmid (Addgene plasmid 1895011 ) was obtained for generating HIF2A mutants. Site-directed mutagenesis was accomplished using the Quikchange Lightning SiteDirected Mutagenesis Kit (Agilent Technologies, Santa Clara, CA).
Hydroxylation assay
An HIF hydroxylation assay was performed as described previously.2 Briefly, synthetic HIF-2α peptides were incubated with recombinant PHD2. HIF-2α peptides were then purified by using Streptavidin Magnetic Beads (Pierce, Rockford, IL). Prolyl hydroxylation of the peptides was then analyzed by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry.
Binding assay
Recombinant VHL protein was translated in a cell-free system in the presence of [35S]-methionine (VHL IVT). Radioactive labeled VHL protein was incubated with recombinant PHD2 and synthetic HIF-2α peptides. Bound VHL protein was eluted and the quantity was determined through scintillation counting and autoradiography.
Chromatin immunoprecipitation assay
HIF-2α recombinants were transfected to HeLa cells for 72 hours. A chromatin immunoprecipitation assay was performed using a SimpleChIP Plus Enzymatic Chromatin IP Kit (Cell Signaling Technology, Danvers, MA). An anti-HA tag antibody (OriGene, Rockville, MD) was used for immunoprecipitation. Precipitated DNA was analyzed by quantitative real-time polymerase chain reaction measurement of the divalent metal transporter 1 (DMT1) promoter containing hypoxia response elements. To analyze ubiquitination, HIF-2α–transfected cells were incubated with MG-132 to accumulate ubiquitinated protein. HIF-2α protein was precipitated with antibody against HA-tag and probed with anti-ubiquitin primary antibody (Abcam, Cambridge, MA).
Immunohistochemistry staining
Immunohistochemistry staining was performed as previously described.12 Formalin-fixed, paraffin-embedded tissue sections were probed with HIF-2α primary antibody (Novus Biologicals, Littleton, CO) and 3,3′-diaminobenzidine staining. Slides were visualized using a Leica DM LB light microscope with a magnification of ×40.
Cycloheximide chase assay
Wild-type HIF-2α or ΔHIF-2α-L529P recombinants were transfected into HeLa cells for 72 hours and exposed to cycloheximide (CHX) to halt protein synthesis. HIF-2α protein residue was determined by western blot.
Structural and energetic modeling
PDB structure ID 3HQR was used as a template, because the carboxy-domain terminal oxygen-dependent degradation domain is conserved between HIF-1α and HIF-2α. Mutagenesis, rotamer selection, and visualization of the appropriate peptide were accomplished using PyMol. Electrostatic interactions were calculated from a Poisson-Boltzmann model using the program APBS.
Results and discussion
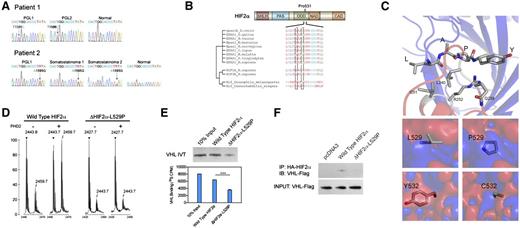
Recently, we identified the first somatic mutations in HIF2A in 2 patients with either multiple paragangliomas or somatostatinomas together with polycythemia.13 Herein, we present 2 more patients with similar genetic changes and clinical manifestations secondary to HIF2A mutations. These 2 female patients presented with polycythemia (at ages 5 and 7 years) followed by the development of multiple abdominal paragangliomas or somatostatinomas. All paragangliomas in these patients presented with a typical noradrenergic biochemical phenotype. No family history of polycythemia, paraganglioma, or somatostatinoma was found in any of these patients. The 2 new unrelated patients presented in this report were found to have 2 novel mutations of HIF2A in tumors cells [c. 1586T>C (p. L529P) and c. 1595A>G (p. Y532C), Figure 1A]. The mutations were identical in multiple tumors from each affected individual, but not in DNA from adjacent normal tissue. Alignments of multiple HIF-2α peptide sequences demonstrated that L529 and Y532 are located in the vicinity of the primary hydroxylation site of HIF-2α protein (Figure 1B). Structurally, these mutations involve residues with key hydrophobic and hydrogen bond interactions with PHD2. Using an analogous crystal structure involving the ODD domain of HIF-1α and the catalytically vital β2β3/loop region of PHD2, we found that these mutations cause local electrostatic changes that disrupt the ODD domain-PHD2 interaction (Figure 1C). We then investigated functional changes in the mutant HIF-2α proteins.
Novel HIF2A mutations disrupt cellular oxygen sensing. (A) Mutation analysis showing heterozygous mutation of HIF2A gene in tumor specimen but not adjacent normal tissue. (B) Peptide sequence alignment of prolyl hydroxylation domain of HIF-2α. (C) Electrostatic changes of single-site mutations in PHD2-HIF-α complex. Interaction between leucine (L) and tyrosine (Y) and PHD2 residues (top) and electrostatic changes resulting from L529P and Y532C substitution (bottom; red, negative; blue, positive; ±5 kT/e) (D) Peptide hydroxylation assay through matrix-assisted laser desorption ionization-time-of-flight showing absent hydroxylation in ΔHIF-2α-L529P mutant. (E) Peptide binding assay of radioactive labeled VHL and HIF-2α-derived peptides, showing reduced VHL binding in HIF-2α mutant. (F) Immunoprecipitation assay showing decreased VHL binding in HIF-2α mutant.
Novel HIF2A mutations disrupt cellular oxygen sensing. (A) Mutation analysis showing heterozygous mutation of HIF2A gene in tumor specimen but not adjacent normal tissue. (B) Peptide sequence alignment of prolyl hydroxylation domain of HIF-2α. (C) Electrostatic changes of single-site mutations in PHD2-HIF-α complex. Interaction between leucine (L) and tyrosine (Y) and PHD2 residues (top) and electrostatic changes resulting from L529P and Y532C substitution (bottom; red, negative; blue, positive; ±5 kT/e) (D) Peptide hydroxylation assay through matrix-assisted laser desorption ionization-time-of-flight showing absent hydroxylation in ΔHIF-2α-L529P mutant. (E) Peptide binding assay of radioactive labeled VHL and HIF-2α-derived peptides, showing reduced VHL binding in HIF-2α mutant. (F) Immunoprecipitation assay showing decreased VHL binding in HIF-2α mutant.
To determine the functional effect of these 2 novel HIF-2α mutations, we first performed an in vitro hydroxylation assay as previously described.14 Consistent with previous reports, wild-type HIF-2α peptides were hydroxylated with PHD2, as shown by the increased mass-to-charge ratio (Figure 1D). In contrast, no obvious hydroxylation of ΔHIF-2α-L529P peptide was seen within the same assay. Additionally, an in vitro binding assay was performed to investigate the role of the L529P mutation on VHL protein binding, a key step in regulation. Wild-type VHL protein binds to wild-type HIF-2α peptides hydroxylated by PHD2. However, the binding efficiency reduced to 55.6% in HIF-2α peptides with L529P amino acid substitution (Figure 1E). The change in VHL binding affinity was confirmed through an immunoprecipitation assay. HA-HIF-2α or ΔHIF-2α-L529P plasmids were cotransfected with VHL-Flag vector in HeLa cells. Total protein was extracted and precipitated with antibody against the HA-tag. VHL binding was determined through western blot of VHL-Flag. We identified VHL binding to wild-type HIF-2α, whereas the binding was reduced to 32.4% in ΔHIF-2α-L529P protein (Figure 1F).
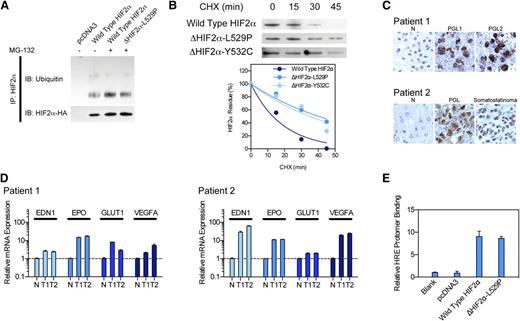
To investigate the stability of HIF-2α protein, we performed an immunoprecipitation assay to measure HIF-2α ubiquitination. We found that the wild-type HIF-2α protein was extensively ubiquitinated, whereas the ubiquitination of the ΔHIF-2α-L529P mutant was reduced to 62.9% (Figure 2A). We further confirmed the impact of the HIF-2α mutation by assessing stability through a CHX assay. Consistent with previous reports, the HIF-2α protein was extremely unstable and degraded shortly upon CHX treatment, with a half-life of 13.1 minutes. In contrast, ΔHIF-2α-L529P and ΔHIF-2α-Y532C mutants exhibited a longer protein lifespan, with a half-life of 40.2 and 34.84 minutes, respectively (Figure 2B). We confirmed HIF-2α stabilization through identifying robust HIF-2α expression, but not HIF-1α, by immunohistochemistry in tumor specimens (Figure 2C). Finally, we confirmed changes in HIF-2α function by quantitative real-time polymerase chain reaction. We identified increased mRNA levels of hypoxia-related genes such as EDN1, EPO, GLUT1, and VEGFA (Figure 2D). A chromatin immunoprecipitation assay also confirmed that the DNA-binding affinity of ΔHIF-2α-L529P remains intact when compared with wild-type HIF-2α (Figure 2E).
HIF2A mutation lead to pseudohypoxic change and tumor formation. (A) Immunoprecipitation assay measuring HIF-2α ubiquitination, showing decreased protein ubiquitination in mutant protein. (B) CHX assay measuring HIF-2α stability, showing elongated protein half-life. (C) Immunohistochemistry staining of HIF-2α in tumor specimens, showing increased HIF-2α protein in tumors. (D) Quantitative polymerase chain reaction assay showing increased hypoxia-related gene expression in tumor specimen. (E) Chromatin immunoprecipitation assay showing intact DNA-binding efficiency of mutant HIF-2α.
HIF2A mutation lead to pseudohypoxic change and tumor formation. (A) Immunoprecipitation assay measuring HIF-2α ubiquitination, showing decreased protein ubiquitination in mutant protein. (B) CHX assay measuring HIF-2α stability, showing elongated protein half-life. (C) Immunohistochemistry staining of HIF-2α in tumor specimens, showing increased HIF-2α protein in tumors. (D) Quantitative polymerase chain reaction assay showing increased hypoxia-related gene expression in tumor specimen. (E) Chromatin immunoprecipitation assay showing intact DNA-binding efficiency of mutant HIF-2α.
In summary, we have identified 2 additional somatic mutations in patients presenting with a cluster of paraganglioma, somatostatinoma, and polycythemia. These mutations appear to disrupt the prolyl hydroxylation domain of HIF-2α, abolish the modification by PHDs, and, as a result a decrease in VHL recognition and VHL-associated degradation. Once translocated to the nucleus, HIF-2α leads to increased expression of hypoxia-related genes. The finding of additional HIF2A mutations in tumors with the same disease cluster implicates HIF2A is a causative gene for this specific combination of diseases.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients and their families for their participation and James W. Nagle for help with DNA sequencing.
This work was supported by the Intramural Research Program of the National Institute of Neurological Disorders and Stroke and the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Authorship
Contribution: C.Y., K.P., and Z.Z. designed research; C.Y., M.G.S., J.M., T.T.H., and S.R. performed research; C.Y., M.G.S., J.M., K.P., and S.R. collected data; C.Y., S.R., J.T.P., R. Lechan, R. Lonser, K.P., and Z.Z. analyzed and interpreted data; and C.Y., S.R., K.P., and Z.Z. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Zhenping Zhuang, National Institute of Neurological Disorders and Stroke, National Institutes of Health, 9000 Rockville Pike, Building 10 / Room 7N246, Bethesda, MA 20894; e-mail: zhuangp@ninds.nih.gov; and Karel Pacak; e-mail: karel@mail.nih.gov.
References
Author notes
C.Y., K.P., and Z.Z. contributed equally to this study.