Key Points
PA28g acts as a co-repressor of HTLV-1 p30 to suppress virus replication and is required for the maintenance of viral latency.
HTLV-1 has evolved a unique function mediated by its posttranscriptional repressor p30, which is not found in HTLV-2.
Abstract
The establishment of a latent reservoir by human tumor viruses is a vital step in initiating cellular transformation and represents a major shortcoming to current therapeutic strategies and the ability to eradicate virus-infected cells. Human T-cell leukemia virus type 1 (HTLV-1) establishes a lifelong infection and is linked to adult T-cell leukemia lymphoma (ATLL). Here, we demonstrate that HTLV-1 p30 recruits the cellular proteasome activator PA28γ onto the viral tax/rex mRNA to prevent its nuclear export and suppress virus replication. Interaction of p30 with a PA28γ retaining fully functional proteasome activity is required for p30's ability to repress HTLV-1. Consistently, HTLV-1 molecular clones replicate better and produce more virus particles in PA28γ-deficient cells. These results define a unique and novel role for the cellular factor PA28γ in the control of nuclear RNA trafficking and HTLV-1–induced latency. Importantly, knockdown of PA28γ expression in ATLL cells latently infected with HTLV-1 reactivates expression of viral tax/rex RNA and the Tax protein. Because Tax is the most immunogenic viral antigen and triggers strong CTL responses, our results suggest that PA28γ-targeted therapy may reactivate virus expression from latently infected cells and allow their eradication from the host.
Introduction
The establishment of a latent reservoir by human tumor viruses is a vital step in initiating cellular transformation and represents a major shortcoming to current therapeutic strategies and the ability to eradicate virus-infected cells. Human tumor viruses establish persistent infections and latent reservoirs in their host, eventually leading to the emergence of transformed cancer cells.1 Due to the oncogenic potential associated with persistent infection of human tumor viruses, development of therapeutic vaccines has been the focus of intense research. Breaking virus latency to force virus expression and the simultaneous use of antiviral drugs to prevent de novo infection is an attractive therapeutic option to unmask and expose infected cells to a patient's immune system.
Human T-cell leukemia virus type 1 (HTLV-1) infection is associated with the development of adult T-cell leukemia lymphoma (ATLL).2-4 The low incidence and the long latency of HTLV-1–associated ATLL suggest that, in addition to viral infection, accumulation of genetic and epigenetic defects are required for cellular transformation and disease progression in vivo. HTLV-1 virus particles are poorly infectious, and HTLV-1 antigens elicit vigorous humoral and cell-mediated immune responses and present very low antigenic variability.5 Thus, reducing expression of viral antigens is essential in virus subsistence in an infected host. The existence of long-lived infected cells must result from proliferation of latently infected cells coupled with the maintenance of a latent reservoir to compensate for the loss of infected cells after virus activation. Because HTLV-1 infection is associated with the development of 2 diseases (ATLL and HTLV-associated myelopathy/tropical spastic paraparesis [HAM/TSP]) with fundamental differences in virus-host interaction, virus replication and pathogenesis, there has been confusion regarding the establishment or not of a latent reservoir by HTLV-1 in vivo. However, it is clear that a latent reservoir infected with HTLV-1 does exist in vivo because the existence of infected T-cell clones with the same provirus integration sites can be found at several years intervals in many ATLL patients.6-8 It is unclear if these infected clones are represented by a distinct T-cell population or by less differentiated hematopoietic precursors.
We previously found that HTLV-1 encodes a potent negative regulator of its own expression and replication.9 The doubly spliced p30 encodes a small basic nuclear/nucleolar protein that specifically interacts with tax/rex viral RNA and prevents its nuclear export, thereby reducing the expression of virus positive regulators Tax and Rex.9 Several studies have demonstrated that p30 is able to suppress virus replication at physiologic levels when expressed in the context of an HTLV-1 molecular clone. Interestingly, HBZ RNA was recently found to act as an anti-sense for p30 RNA and to promote Tax expression.10 Expression of p30 is essential for virus replication in dendritic cells and for virus spread and establishment of a persistent infection in nonhuman primates.11
Although the use of viral proteins has paved the way to our current understanding of the cellular machinery involved in nuclear export of RNA,12 the role of the cellular factors involved in nuclear retention of RNA is still poorly understood. Proteasome activator PA28γ (also known as REGγ, PSME3, or Ki Antigen) has been shown to control the formation of nuclear speckles, a subdomain involved in pre-mRNA processing.13 Although PA28α and PA28β are involved in the processing of intracellular antigens, the role of PA28γ has remained elusive.
In this study, we identified PA28γ as a cellular factor required for HTLV-1 p30 to inhibit virus replication. Using RNA in situ hybridization and RIP (RNA-immunoprecipitation) assays, we found that p30 recruits PA28γ onto viral tax/rex mRNAs, leading to their retention in the nucleus. We show that several HTLV-1 infectious molecular clones replicate more efficiently and produce more virus particles when transferred into PA28γ knockdown cells. The cooperative effect of p30 and PA28γ enables HTLV-1 to conceal itself and avoid immune surveillance at key stages of infection. Our results further demonstrated that knocking down PA28γ expression allowed reactivation of viral gene expression in ATLL cells latently infected with HTLV-1. These results uncover a novel function for the cellular factor PA28γ in inhibiting viral mRNA nuclear export and suppressing replication of the human retrovirus HTLV-1.
Methods
Cell lines and transfections
PA28γ−/− mouse embryo fibroblasts (MEFs) were generously provided by Dr Lance Barton (Austin, TX). Other cell lines, including 293T and HeLa, were obtained from the ATCC. 293-FT cells were transfected by calcium phosphate (Invitrogen) or polyfect (QIAGEN). MEF cells were transfected using lipofectamine (Invitrogen). 293T derived FT-27 were generated by infection with high titer PA28γ shRNA lentivirus (OriGene) followed by puromycin selection. ATLL-derived cells latently infected with HTLV-1, MT1, and TL-Om1 were provided by Dr M. Maeda (University Sakyo-ku). Acute ATL samples were previously reported.14
Plasmids and cloning of PA28γ mutants
PA28α cDNA was kindly provided by Dr Klaus Fruh (Oregon Health & Science University), and was subcloned with an N-terminal Flag-tag into pcDNA3.1. HTLV-2 p28 was subcloned into pMH3xHA. p30(67-241) were generated by PCR and cloned into pMH3xHA. PA28γ (G150S), PA28γ (N151S), PA28γ (N151Y), PA28γ (R186C), PA28γ (D210V), and PA28γ (P245A) were generated by site-directed mutagenesis (Stratagene). PA28γ (Δ71-102) was generated by overlapping PCR. PA28γ (α20-25) was constructed by replacing amino acids 21-26 from PA28γ (RITSEA) with amino acids 20-25 from PA28α (DLCTKT). All mutants were verified by sequencing. HTLV-1 molecular clones ACH, pBST, X1MT, and pABΔp30 were previously described.
Western blot, immunoprecipitations, and ELISA
The following antibodies were used: anti–HA-HRP (3F10; Roche), anti-FLAG (M2; Sigma-Aldrich), anti-PA28γ (BioMol), anti-tubulin (Santa Cruz), and anti–mouse-HRP and anti–rabbit-HRP secondary antibodies (Santa Cruz). Proteins were immunoprecipitated with either anti-FLAG (M2; Sigma-Aldrich), anti-HA 12CA5 (Roche), or anti-PA28γ (BioMol). Detection of intracellular HTLV-1 p24 and virus production was performed by ELISA.
Immunofluorescence
Transfected cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton-X-100, blocked with 0.1% FBS/PBS and stained with primary antibodies at room temperature for 90 minutes. Cells were stained with secondary antibodies, anti–mouse Alexa488, or anti–mouse Alexa546 (Molecular Probes), at room temperature for 1 hour, and stained with DAPI before mounting. Cells were visualized using a Nikon Eclipse Ti-S inverted fluorescent microscope.
Purification of p30 and mass spectrometry analysis
293T cells were transfected with pMH p30, and cells were washed twice with PBS and lysed in 1% NP-40 lysis buffer. p30 was immunoprecipitated with anti-HA (3F10), and proteins were analyzed by SDS-PAGE and stained with Coomassie brilliant blue. Protein bands were excised from the gel and analyzed by mass spectrometry at the University of Kansas Proteomics Facility (Lawrence, Kansas).
Elution of PA28γ complexes and proteasome assays
To elute PA28γ-containing complexes, after immunoprecipitation of Flag-PA28γ complexes, samples were incubated with 100 ug/mL FLAG peptide (Sigma-Aldrich). Proteasome activity assays were performed as described.15 Increasing amounts of protein samples were incubated with purified MCP in the presence of either sLLVY or LRR for 20 minutes.
RNA immunoprecipitation assays
FT-27 cells were transfected by calcium phosphate (Invitrogen) with PBST, p30HA, and/or Flag-REGγ. After 48 hours, cells were washed in PBS and lysed in NP-40 lysis buffer with ribonucleoside vanadyl complexes (Sigma-Aldrich), RNaseOUT (Invitrogen), phosphatase, and proteases inhibitors. Immunoprecipitations were carried out overnight with anti-FLAG antibody. Immunoprecipitates were lysed in TRIzol (Invitrogen) for RNA extraction. RNA normalized to PA28γ expression was used for RT-PCR using the OneStep RT-PCR kit (QIAGEN) with tax/rex and p21rex specific primers. Equal amounts of RNA were subject to PCR, without reverse transcription, to control for potential DNA contamination. Gel-extracted RT-PCR products were cloned into the pGEM-T Easy vector (Promega) and sequenced.
RNA FISH for detection of the tax/rex RNA
Cells grown on glass coverslips were transfected with GFP-p30 and an HTLV-1 molecular clone, fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100 in PBS on ice for 5 minutes and washed twice with PBS and with 2 × SSC-50% formamide for 5 minutes. The hybridization mixture (30 ng of FISH DNA probes against target tax/rex RNA directly labeled with Cy3 by nick translation and 40 μg yeast tRNA per coverslip) was made to a final concentration of 2 × SSC, 20% deionized formamide, 10% dextran sulfate, 2mM vanadyl-ribonucleoside complex and 0.02% RNase-free BSA. A hybridization mixture (40 μL) was placed onto a coverslip sealed to microscopic slides by rubber cement and incubated in a chamber moistened with 2 × SSC overnight at 37°C. Coverslips were rinsed twice for 30 minutes with 2 × SSC-50% formamide at 37°C and for 5 minutes with PBS at room temperature. At this point, the RNA FISH protocol was combined with the indirect immunofluorescence using specific primary antibodies against PA28γ. Cells were observed on a Zeiss LSM510 confocal microscope using a 100 × 1.4 N.A. Plan Apochromat oil objective. Images were collected as vertical z stacks (z; 200 nm) and projected as maximum intensity projections.
Real-time quantitative RT-PCR
Total RNA was prepared using TRIzol reagent (Invitrogen). After DNAse I treatment, the RNA was reverse transcribed and the cDNA was used for real-time PCR. The PCR primers RPX3 5′-ATCCCGTGGAGACTCCTCAA-3′ and RPX4 5′-AACACGTAGACTGGGTATCC-3′ were used for tax/rex RNA. Real time RT-PCR was performed using RT2 SYBR Green qPCR Mastermix (SA Bioscience) with the following primers: LTR2, tax/rexR (5′-GGAAGTGGGCCATGGTG-3′), p30 splice specific TF4 (5′-CAGCATAGTGCCATGGTG-3′), GADPH F: (5′-GAAGGTGAAGGTCGGAGTC-3′), and GADPH R: (5′-GAAGATGGTGATGGGATTTC-3′). The expression of tax/rex was normalized to the GAPDH. Taqman real time gene expression assays were run on an ABI StepOnePlus system (Applied Biosystems) and data were analyzed with StepOnePlus Version 2.0 software. Standard curves were generated using 10-fold serial dilutions of a double standard plasmid containing the target regions of Tax/Rex and GAPDH genes. The Tax/Rex copy number was reported as Tax/Rex copy number/106GAPDH copy number. RPX3 and RPX4 primers were used for Tax/Rex. The Taqman probe for Tax/Rex (5′-FAM-CCACCAACACCATGGCCCACTTCC-IBFQ-3′) is splice-site spanning. The GAPDH primers and Taqman probe for GAPDH are from the Taqman GAPDH control kit (Applied Biosystems).
Tax intracellular staining
Control and PA28γ knockdown MT-1 and TL-Om1 cells were fixed with 2% paraformaldehyde in PBS for 20 minutes on ice, washed, then permeabilized with 0.1% Triton X-100 in PBS for 5 minutes on ice. Cells were washed and a primary anti-Tax antibody was added in 7%FBS/PBS for approximately 2 ½ hours at room temperature. Alternatively, unstained cells were incubated in 7% FBS/PBS without a Tax antibody. All cells were then washed and incubated with FITC Donkey anti–mouse 488 (Santa Cruz) in 7% FBS/PBS for 35 minutes at room temperature. Cells were subsequently washed and resuspended in PBS for FACS analysis.
Results
HTLV-1 p30 forms specific complexes with the proteasome activator subunit PA28γ
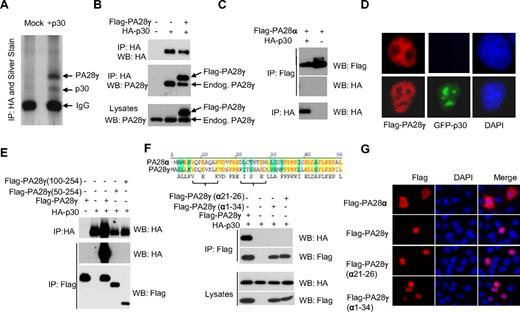
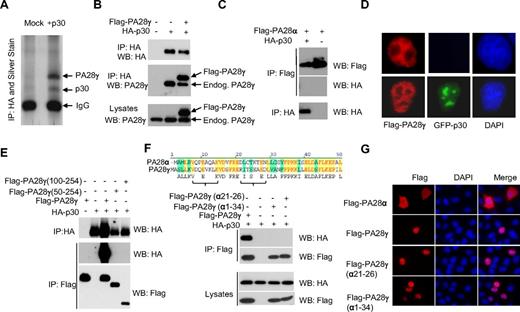
Using large-scale immunoprecipitation of HA-tagged p30 and mass spectrometry, we identified the nuclear proteasome activator PA28γ as a specific cellular interacting partner of p30 (Figure 1A). Additional potential p30-interacting partners identified were protein arginine methyltransferase 5 (PRMT5), tubulin α-1B chain (TBA1B) and importin-5 (IPO5). Specific interaction between p30 and PA28γ was further validated by coimmunoprecipitation of HA-p30 and FLAG-PA28γ, as well as p30 interaction with endogenous PA28γ (Figure 1B). To gain insights into the region of p30 involved in binding to PA28γ, we generated several N-terminal deletion mutants. We identified that a deletion of 67 amino acid residues from the N-terminus of p30 ablated the ability of p30 to interact with PA28γ (data not shown). We confirmed that p30 and PA28γ form a complex in vivo as shown by their coelution in a 200-kDa complex using gel filtration assays (data not shown). As expected from the above data, NΔ67p30 did not coelute with PA28γ. Although p30 and Rex have been shown to interact with each other,16 we did not detect any Rex binding to PA28γ and found that the p30/ PA28γ complex occurs independently of Rex (data not shown). The crystal structure of PA28α has been reported and consists of a series of 4 parallel helices, and considering the degree of homology, PA28γ is predicted to possess the same overall structure as PA28α.17 Therefore, we further investigated whether p30 might bind to the PA28γ substrate recognition site. We deleted the region proposed to play a critical role in substrate recognition,18 PA28γΔ71-103, but this mutant still interacted with p30 (data not shown). We then constructed 2 additional PA28γ N-terminal deletion mutants lacking 50 and 100 amino acids (PA28γ 100-254 or PA28γ 50-254) and tested their interaction with p30. Neither of these mutants was able to bind to p30 (Figure 1E). These results indicate that the p30 binding domain probably resides within the first 50 amino acid residues of PA28γ. Because no interaction between p30 and PA28α was detected (Figure 1C), we searched for regions of dissimilarity between PA28α and PA28γ (Figure 1F) and generated a fusion of PA28α(1-32) and PA28γ (34-254). Binding assays revealed that this mutant was unable to bind to p30 (Figure 1F). Based on matched peptides representation identified by mass spectrometry analyses within this region, we swapped amino acids from PA28α to PA28γ, and generated a mutant PA28γ (α21-26), which completely lost the ability to bind to p30 (Figure 1F). The lack of interaction between p30 and PA28γ (α21-26) was not the result of mislocalization because this mutant shared PA28γ localization in the nucleus when analyzed by immunofluorescence (Figure 1G).
The proteasome activator PA28γ is a cellular partner for HTLV-1 p30. (A) 293T cells transfected with HA-p30 were immunoprecipitated with α-HA (3F10), and samples were analyzed by SDS-PAGE and silver staining. Specific bands were analyzed by mass spectrometry. (B) 293T cells were co-transfected with HA-p30 and Flag-PA28γ, and cell lysates were immunoprecipitated with the indicated antibodies and blotted with either HA or PA28γ antibodies to detect both FLAG-tagged and endogenous PA28γ. (C) HTLV-1 p30 does not interact with PA28α. 293T cells were transfected, immunoprecipitated, and immunoblotted with the indicated antibodies. (D) p30 expression does not alter PA28γ immunolocalization. Cells were transfected with GFP-p30 with or without FLAG-PA28γ. Immunofluorescence was performed after 48 hours. (E) p30 interacts within the first 50 amino acid residues of PA28γ. 293T cells were co-transfected with the indicated PA28γ mutant constructs along with p30 HA, and were immunoprecipitated and immunoblotted. (F) Alignment of the PA28γ and PA28α amino acid sequence. Swapping a 6 amino acid α helix region from PA28γ with those of PA28α abrogates p30 interaction with PA28γ in cotransfection experiments. (G) Nuclear localization of PA28γ(α21-26) is not altered. HeLa cells were transfected with PA28α, PA28γ, PA28γ(α21-26), and PA28γ(α1-34), fixed and stained with α-FLAG, and visualized by immunofluorescence.
The proteasome activator PA28γ is a cellular partner for HTLV-1 p30. (A) 293T cells transfected with HA-p30 were immunoprecipitated with α-HA (3F10), and samples were analyzed by SDS-PAGE and silver staining. Specific bands were analyzed by mass spectrometry. (B) 293T cells were co-transfected with HA-p30 and Flag-PA28γ, and cell lysates were immunoprecipitated with the indicated antibodies and blotted with either HA or PA28γ antibodies to detect both FLAG-tagged and endogenous PA28γ. (C) HTLV-1 p30 does not interact with PA28α. 293T cells were transfected, immunoprecipitated, and immunoblotted with the indicated antibodies. (D) p30 expression does not alter PA28γ immunolocalization. Cells were transfected with GFP-p30 with or without FLAG-PA28γ. Immunofluorescence was performed after 48 hours. (E) p30 interacts within the first 50 amino acid residues of PA28γ. 293T cells were co-transfected with the indicated PA28γ mutant constructs along with p30 HA, and were immunoprecipitated and immunoblotted. (F) Alignment of the PA28γ and PA28α amino acid sequence. Swapping a 6 amino acid α helix region from PA28γ with those of PA28α abrogates p30 interaction with PA28γ in cotransfection experiments. (G) Nuclear localization of PA28γ(α21-26) is not altered. HeLa cells were transfected with PA28α, PA28γ, PA28γ(α21-26), and PA28γ(α1-34), fixed and stained with α-FLAG, and visualized by immunofluorescence.
p30 does not inhibit the intrinsic ability of PA28γ to degrade target substrates through the proteasome
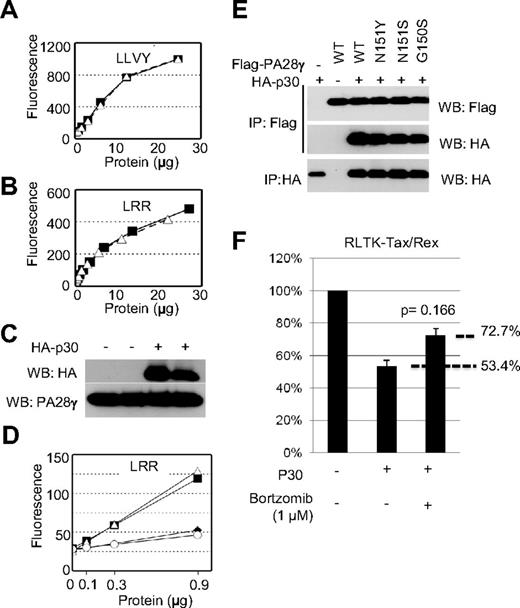
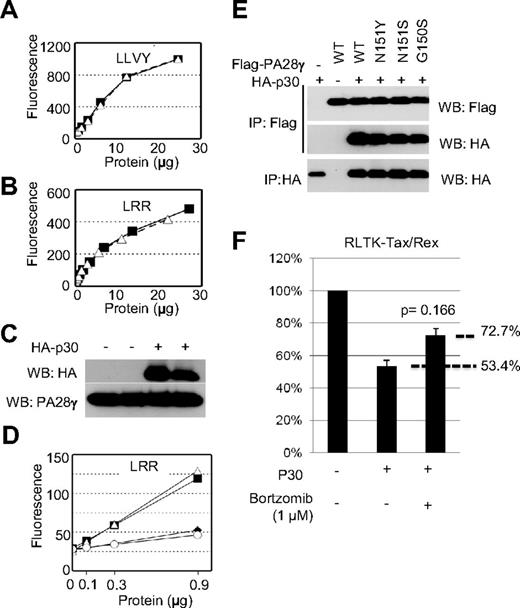
Taking into consideration that p30 can form a complex with PA28γ, we next assessed whether p30 had any effect on the intrinsic proteasomal activity of PA28γ. We generated lysates from cells expressing p30 and tested their ability to degrade fluorogenic peptides in the presence of purified 20S proteasomes. Intriguingly, cell lysates from mock or p30-transfected cells showed no significant difference in the ability to activate the proteasome toward either LLVY or LRR (Figure 2A-B), substrates known to be degraded specifically by the PA28α and PA28γ proteasome activators, respectively.15 Western blot fractions showed equivalent levels of endogenous PA28γ and of transfected p30 in duplicate samples (Figure 2C). To rule out the possible deleterious effects of endogenous 20S proteasomes and activators from our cell lysates in these assays, we also purified PA28γ in the presence or absence of HA-p30. Co-transfected cells were lysed, immunoprecipitated using anti-FLAG antibody, eluted with FLAG-peptide, PA28γ eluates, in the presence or absence of p30, and were then used in the aforementioned proteasome activation assays. As a negative control, we also purified PA28γ (N151S), which was previously shown to be unable to activate the proteasome.18 In fact, mock and PA28γ (N151S) were both unable to induce cleavage of LRR, whereas PA28γ and PA28γ-p30 were both equally capable of inducing LRR cleavage (Figure 2D). Equivalent amounts of protein were included in all assays and verified by Western blot and Coomassie staining of eluted protein samples (data not shown). Altogether, these results demonstrate that p30 does not inhibit the intrinsic ability of PA28γ to degrade target protein substrates through the proteasome.
p30 binding to PA28γ does not inhibit PA28γ proteasomal activity. (A-B) Cell lysates from cells expressing p30 (open triangles) or mock-transfected cells (closed squares) were used in proteasomal activation assays as described in “Methods.” Samples were assayed for their ability to cleave the fluorescent substrates LLVY (A) or LRR (B), which detect the activity of PA28α and PA28γ, respectively. (C) Cell lysates from panel A were Western blotted as indicated. (D) Eluted protein complexes isolated as in panel A were subjected to proteasomal activity assays as described in panel A. Mock-transfected (open circles), PA28γ (closed squares), PA28γ + p30 (open triangles), or PA28γ N151S (closed diamonds) were assayed for their ability to cleave LRR to detect PA28γ activity. (E) The interaction between the PA28γ and PA28γ mutant defective for proteasome activation (N151Y, N151S, G150S), (Δ245-254, P245A) and p30HA were analyzed after transient expression in 293T cells. (F) 293T cells were transfected with RL-TK-tax/rex in the presence or absence of p30 and treated with proteasome inhibitor Bortzomib at 1 micromolar for 24 hours and assayed for renilla activity.
p30 binding to PA28γ does not inhibit PA28γ proteasomal activity. (A-B) Cell lysates from cells expressing p30 (open triangles) or mock-transfected cells (closed squares) were used in proteasomal activation assays as described in “Methods.” Samples were assayed for their ability to cleave the fluorescent substrates LLVY (A) or LRR (B), which detect the activity of PA28α and PA28γ, respectively. (C) Cell lysates from panel A were Western blotted as indicated. (D) Eluted protein complexes isolated as in panel A were subjected to proteasomal activity assays as described in panel A. Mock-transfected (open circles), PA28γ (closed squares), PA28γ + p30 (open triangles), or PA28γ N151S (closed diamonds) were assayed for their ability to cleave LRR to detect PA28γ activity. (E) The interaction between the PA28γ and PA28γ mutant defective for proteasome activation (N151Y, N151S, G150S), (Δ245-254, P245A) and p30HA were analyzed after transient expression in 293T cells. (F) 293T cells were transfected with RL-TK-tax/rex in the presence or absence of p30 and treated with proteasome inhibitor Bortzomib at 1 micromolar for 24 hours and assayed for renilla activity.
Proteasomal activity of PA28γ is required for p30-mediated inhibition of tax/rex RNA
The carboxy-terminus of PA28γ binds to the 20S proteasome core. Amino acids 150-156, which are identical to the corresponding region of PA28α (146-151) make up the active proteolytic site involved in directing substrates into the proteasomal core for degradation.19 To gain additional insights about the physiologic function of the p30-PA28γ interaction, we generated several FLAG-tagged mutants of PA28γ that were previously characterized to be deficient in activating the proteasome, (PA28γ N151Y, N151S, G150S, P245A, and Δ246-254).18 Immunoprecipitation studies indicated that p30 efficiently interacted with all PA28γ proteasome-deficient mutants in human cells (Figure 2E and data not shown). In the absence of a specific inhibitor for PA28γ proteasome activity, we used a general proteasome inhibitor (bortzomib) in our RLTK-tax/rex reporter assay. Our results indicated that inhibition of proteasome activity rescued approximately 40% of p30-mediated inhibition of tax/rex (Figure 2F). Although this experiment does not offer direct proof because it may have off-target effects, it is provided here as general support to our findings. We think that PA28γ-specific inhibitors may exert a more potent effect and research of synthetic small inhibitors is under investigation.
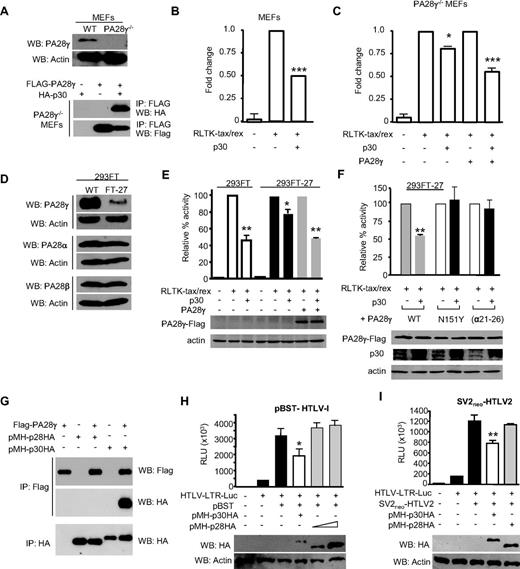
We next demonstrated that p30 interacted with PA28γ when expressed in PA28γ−/− MEFs20 (Figure 3A). We then used an established assay based on the expression of a Renilla-fused tax/rex sequence containing the p30 response element.9 In this assay, p30 expression resulted in a dose-dependent decrease of Renilla activity consistent with the p30-mediated nuclear retention of the tax/rex mRNA. Results from these studies demonstrated that p30 had a reduced effect on inhibition of the tax/rex mRNA retention in PA28γ−/− cells compared with the wild-type MEF cells, but p30 repressive functions were fully restored on transfection of a FLAG-PA28γ expression vector (Figure 3B-C). To prove the importance of our findings in human cells, we used shRNA vectors to knock down endogenous PA28γ expression in human 293FT cells. After selection, FT-27 cells showed a significant decrease in PA28γ expression but had no effect on the related proteins PA28α and PA28β (Figure 3D). We found that p30 was able to efficiently decrease RLTK-tax/rex activity in 293FT cells, but not in FT-27 cells knocked down for endogenous PA28γ (Figure 3E). These effects were specific to the lack of PA28γ expression because exogenous expression of PA28γ fully restored p30-mediated repression of the RLTK-tax/rex (Figure 3E). It also appears that expression of PA28γ leads to higher expression of p30, although the mechanism involved is unclear at this time. We think that the remaining p30 activity seen in 293-FT-27 cells may be related to residual endogenous PA28γ expression, as shRNA does not achieve a complete knockout of expression. The requirement of PA28γ expression for p30-mediated repression of HTLV-1 was confirmed in several 293FT individual clones knocked down for PA28γ as well as in additional 293T and HeLa cell lines knocked down for PA28γ (data not shown). Consistent with the these results and their lack of interactions, p30 was also unable to inhibit tax/rex mRNA export when expressed along with PA28γ (α21-26; Figure 3F). Altogether, these results confirm that p30 requires binding of PA28γ to repress tax/rex mRNA expression. We then tested the requirement of PA28γ proteasomal activity for p30-mediated repression of HTLV-1 replication. Interestingly, p30 was unable to inhibit tax/rex mRNA when the N151Y PA28γ proteasome deficient mutant was used (Figure 3F). Similar results were obtained with additional PA28γ proteasome-deficient mutants (N151Y, N151S, G150S, and P245A; data not shown). Taken together, our findings demonstrate that the p30-PA28γ interaction and PA28γ proteasome activity are both required for the inhibition of tax/rex mRNA expression.
Both interaction with PA28γ and PA28γ proteasomal activity are required for p30-mediated inhibition of tax/rex RNA. (A) p30 retains the ability to interact with exogenously expressed PA28γ in MEF PA28γ knockout cells. (B-C) p30-mediated repression of tax/rex RNA is reduced in MEF PA28γ−/− compared with MEF wild-type cells. p30 activity is rescued by exogenous expression of FLAG-PA28γ. (D) Characterization of FT-27 cells stably knocked-down for endogenous PA28γ. Western blots show drastically reduced PA28γ expression, whereas expression of related PA28α and PA28β were not affected by shRNA used. (E) p30 efficiently inhibits tax/rex mRNA in 293FT cells but not in FT-27 knocked down for PA28γ expression. Lack of p30-mediated repression was rescued by ectopic expression of a PA28γ expression vector. (F) In contrast to PA28γ, a PA28γ mutant defective for proteasome activation N151Y cannot rescue p30-mediated repression of tax/rex RNA. PA28γ (α21-26), unable to bind p30, was not able to rescue p30-mediated repression of tax/rex RNA. Western blots show p30 and PA28γ expression in transfected cells. (G) FLAG-PA28γ was transfected into 293T cells in the presence or absence of p30-HA or p28-HA expression vectors. Cell lysates were used for immunoprecipitations as indicated. (H) Repressive effects of p30 and p28 were analyzed by transfection of 293T cells with HTLV-1 molecular clone pBST, HTLV-1-LTR-luciferase reporter and either p30HA or p28HA as indicated. Western blots were performed to show relative levels of p30 and p28 expression. Results are representative of 3 independent experiments. (I) Repressive effects of p30 and p28 were analyzed by transfection of 293T cells with HTLV-2 molecular clone SV2neoHTLV-2, HTLV-1-LTR-luciferase reporter and either p30HA or p28HA as indicated. Western blots were performed to show relative levels of p30 and p28 expression. Results are representative of 3 independent experiments, P values for all the panels using 1-way ANOVA with Bonferroni multiple comparison posttest (*P < .5, **P < .01, and ***P < .001).
Both interaction with PA28γ and PA28γ proteasomal activity are required for p30-mediated inhibition of tax/rex RNA. (A) p30 retains the ability to interact with exogenously expressed PA28γ in MEF PA28γ knockout cells. (B-C) p30-mediated repression of tax/rex RNA is reduced in MEF PA28γ−/− compared with MEF wild-type cells. p30 activity is rescued by exogenous expression of FLAG-PA28γ. (D) Characterization of FT-27 cells stably knocked-down for endogenous PA28γ. Western blots show drastically reduced PA28γ expression, whereas expression of related PA28α and PA28β were not affected by shRNA used. (E) p30 efficiently inhibits tax/rex mRNA in 293FT cells but not in FT-27 knocked down for PA28γ expression. Lack of p30-mediated repression was rescued by ectopic expression of a PA28γ expression vector. (F) In contrast to PA28γ, a PA28γ mutant defective for proteasome activation N151Y cannot rescue p30-mediated repression of tax/rex RNA. PA28γ (α21-26), unable to bind p30, was not able to rescue p30-mediated repression of tax/rex RNA. Western blots show p30 and PA28γ expression in transfected cells. (G) FLAG-PA28γ was transfected into 293T cells in the presence or absence of p30-HA or p28-HA expression vectors. Cell lysates were used for immunoprecipitations as indicated. (H) Repressive effects of p30 and p28 were analyzed by transfection of 293T cells with HTLV-1 molecular clone pBST, HTLV-1-LTR-luciferase reporter and either p30HA or p28HA as indicated. Western blots were performed to show relative levels of p30 and p28 expression. Results are representative of 3 independent experiments. (I) Repressive effects of p30 and p28 were analyzed by transfection of 293T cells with HTLV-2 molecular clone SV2neoHTLV-2, HTLV-1-LTR-luciferase reporter and either p30HA or p28HA as indicated. Western blots were performed to show relative levels of p30 and p28 expression. Results are representative of 3 independent experiments, P values for all the panels using 1-way ANOVA with Bonferroni multiple comparison posttest (*P < .5, **P < .01, and ***P < .001).
HTLV-1 p28 does not interact with PA28γ and does not suppress HTLV-1 or HTLV-2 viral replication
HTLV-2 is nonpathogenic in humans but the reasons for this are not clear. Because HTLV-2 p28 shares only limited homology with the amino terminus of p30 that is responsible for the interactions with PA28γ, we assessed p28 binding to PA28γ. HA-tagged p28 was unable to coprecipitate with PA28γ (Figure 3G), suggesting that HTLV-2 p28 does not bind to PA28γ, and that the interaction between p30 and PA28γ is specific to HTLV-1. Although it was previously reported that HTLV-2 p28 has a similar effect as HTLV-1 p30 on virus replication,21 our experiments could not confirm these data and showed no effect for p28. We then obtained HTLV-2 p28 and an HTLV-2 molecular clone from the investigators of the previously mentioned study and cloned both p30 and p28 with the same tag and into the same vector. Our experiments confirmed that p30 represses replication of both HTLV-1 and HTLV-2 molecular clones, but p28 had no specific effect on either HTLV-1 or HTLV-2 replication, even when p28 was overexpressed 5- to 10-fold higher than p30 (Figure 3H-I). Together our data suggest that HTLV-1 has evolved a unique viral protein p30, not found in HTLV-2, which efficiently suppresses HTLV-1 virus expression. The lack of a p30 homologue in HTLV-2 may limit latency and reduce the lifespan of long-lasting infected cells in vivo, thereby reducing the chance of acquiring genetic alterations and developing a disease. Further studies aimed at understanding the lifespan of HTLV-1 and HTLV-2 infected clones in vivo will help resolve these issues.
p30 specifically recruits PA28γ to sequester viral tax/rex mRNA into the nucleus
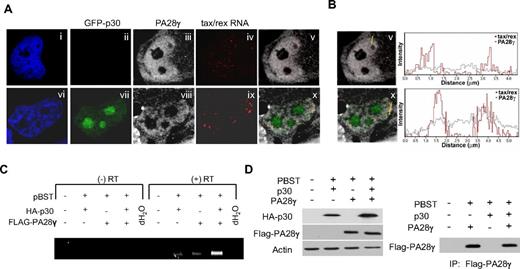
p30 is selectively recruited to the tax/rex mRNA and prevents its nuclear export to the cytoplasm, thereby reducing expression of Tax and Rex, positive regulators of virus expression.11 The molecular mechanism used by p30 to achieve suppression of virus replication has been elusive. Based on the data presented above, we hypothesized that p30 might recruit PA28γ onto the tax/rex RNA and trigger PA28γ-mediated degradation of cellular components of the RNA export machinery, thereby sequestering the viral mRNA in the nucleus. To test this hypothesis, we used RNA in situ hybridization for simultaneous detection of tax/rex mRNA with immunolocalization of PA28γ and p30 proteins in the cells. Our results clearly demonstrated a nuclear accumulation of the tax/rex mRNA in larger foci (Figure 4Ai-x) that colocalized with PA28γ and occurred only in the presence of p30 (Figure 4A). Quantifications of tax/rex RNA and PA28γ colocalization are shown in Figure 4B. These results demonstrate for the first time p30-mediated nuclear retention of the tax/rex RNA in vivo. To further extend these results and test whether PA28γ may be recruited to the tax/rex mRNA we performed RIP assays. Human FT-27 PA28γ knockdown cells were co-transfected with an HTLV-1 molecular clone and a FLAG-PA28γ expression vector in the presence or absence of p30-HA. After immunoprecipitation with an anti-FLAG antibody, PA28γ-bound RNAs were extracted and analyzed by RT-PCR. Results from these assays indicated that PA28γ is specifically recruited to the tax/rex mRNA only when expressed in the presence of p30 (Figure 4C). Similar amounts of PA28γ were expressed in the presence or absence of p30 (Figure 4D) and the identity of the tax/rex mRNA was confirmed by sequencing. Although the primers used in our assays can amplify both tax/rex and p21rex viral mRNAs (Figure 4C), only tax/rex mRNA was amplified in our RIP experiments. These results are consistent with our previous studies showing that tax/rex but not p21rex RNA contains the p30 responsive element, and that p30 forms a complex specifically with the tax/rex mRNA.9
p30 specifically recruits PA28γ to trap viral tax/rex RNA into the nucleus. (A) Simultaneous in situ hybridization detection of the tax/rex viral mRNA, immunolocalization of endogenous PA28γ and GFP-p30 show nuclear accumulation of tax/rex mRNA in the presence of GFP-p30. (B) To quantify the level of colocalization between tax/rex RNA detected by RNA FISH and PA28gamma, we measured their signal intensity profiles in cells with and without p30. The signal intensity profiles indicate that there is an increased level of PA28gamma at the sites of tax/rex RNA localization in the presence of the p30 protein in comparison to the cells without p30. (C) RNA immunoprecipitation shows specific recruitment of PA28γ onto the tax/rex mRNA in the presence of p30. HTLV-1 molecular clone pBST was transfected along with p30HA and/or FLAG-PA28γ into FT-27 PA28γ knockdown cells. Lysates were immunoprecipitated with anti-FLAG antibody and RNA extracted TRIzol and analyzed by RT-PCR for the presence of tax/rex or p21rex mRNA. (D) Western blot was performed on cell lysates to confirm p30HA expression and similar expression of transfected FLAG-PA28γ. Similar amounts of PA28γ were immunoprecipitated using the FLAG antibody as shown by Western blotting of immunoprecipitates.
p30 specifically recruits PA28γ to trap viral tax/rex RNA into the nucleus. (A) Simultaneous in situ hybridization detection of the tax/rex viral mRNA, immunolocalization of endogenous PA28γ and GFP-p30 show nuclear accumulation of tax/rex mRNA in the presence of GFP-p30. (B) To quantify the level of colocalization between tax/rex RNA detected by RNA FISH and PA28gamma, we measured their signal intensity profiles in cells with and without p30. The signal intensity profiles indicate that there is an increased level of PA28gamma at the sites of tax/rex RNA localization in the presence of the p30 protein in comparison to the cells without p30. (C) RNA immunoprecipitation shows specific recruitment of PA28γ onto the tax/rex mRNA in the presence of p30. HTLV-1 molecular clone pBST was transfected along with p30HA and/or FLAG-PA28γ into FT-27 PA28γ knockdown cells. Lysates were immunoprecipitated with anti-FLAG antibody and RNA extracted TRIzol and analyzed by RT-PCR for the presence of tax/rex or p21rex mRNA. (D) Western blot was performed on cell lysates to confirm p30HA expression and similar expression of transfected FLAG-PA28γ. Similar amounts of PA28γ were immunoprecipitated using the FLAG antibody as shown by Western blotting of immunoprecipitates.
Interaction between p30 and PA28γ suppresses replication and virus production of infectious HTLV-1 molecular clones.
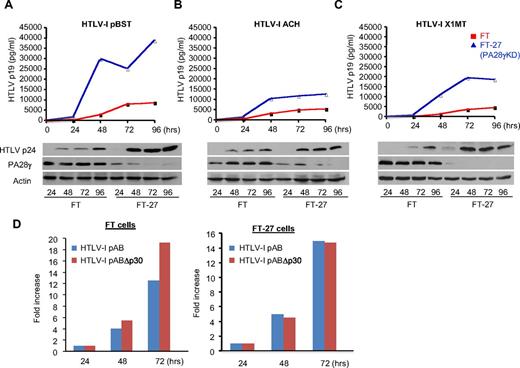
We validated our findings in a physiologic context of viral replication with endogenous expression of both p30 and PA28γ. We hypothesized that p30 should not affect tax/rex mRNA in PA28γ knockdown cells and, consequently, that HTLV-1 should replicate more efficiently in these cells. To test this hypothesis, we used 3 different HTLV-1 molecular clones. Replication of these molecular clones, pBST, ACH, and X1MT,22-24 was monitored by intra-cellular Gag protein expression and extra-cellular virus production measured by ELISA. Our results confirmed that all 3 HTLV-1 molecular clones replicated more efficiently and produced more virus particles when expressed in PA28γ knockdown cells (Figure 5A-C). We further investigated replication of a p30-deleted HTLV-1 molecular clone (pABΔp30) compared with its wild-type counterpart pAB.11 As expected from our previous data and other studies, deletion of p30 prevents the inhibition of positive regulators Tax and Rex and leads to increased virus replication in normal FT cells (Figure 5D). However, there was no significant difference in virus replication between wild-type or p30-deleted HTLV-1 molecular clones in PA28γ-deficient cells. (Figure 5D). Importantly, these results confirmed inhibition of HTLV-1 replication by p30 and PA28γ expressed under natural physiologic conditions.
Inhibition of PA28γ expression increases HTLV-1 replication and virus production. (A-C) ELISA p19 assay was performed every 24 hours for 96 hours after transfection of 293FT and FT-27 cells with distinct HTLV-1 molecular clones (pBST, ACH, and X1MT). Western blots analyses confirmed increased p24 in FT-27 cells lacking PA28γ expression. (D) Replication of HTLV-1 molecular clone pAB and its counterpart pABΔp30 was analyzed in FT and FT-27 cells transfected with respective molecular clones and HTLV-1-LTR-luciferase.
Inhibition of PA28γ expression increases HTLV-1 replication and virus production. (A-C) ELISA p19 assay was performed every 24 hours for 96 hours after transfection of 293FT and FT-27 cells with distinct HTLV-1 molecular clones (pBST, ACH, and X1MT). Western blots analyses confirmed increased p24 in FT-27 cells lacking PA28γ expression. (D) Replication of HTLV-1 molecular clone pAB and its counterpart pABΔp30 was analyzed in FT and FT-27 cells transfected with respective molecular clones and HTLV-1-LTR-luciferase.
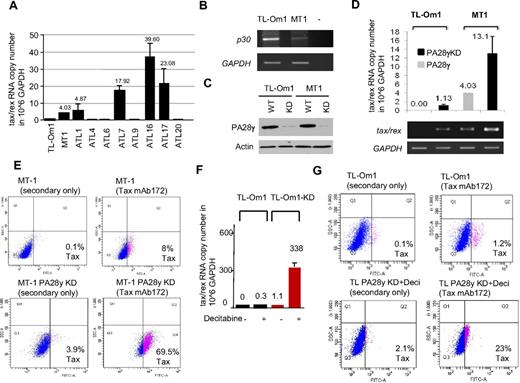
Inhibition of PA28γ expression reactivates HTLV-1 expression from ATLL latently infected human T cells
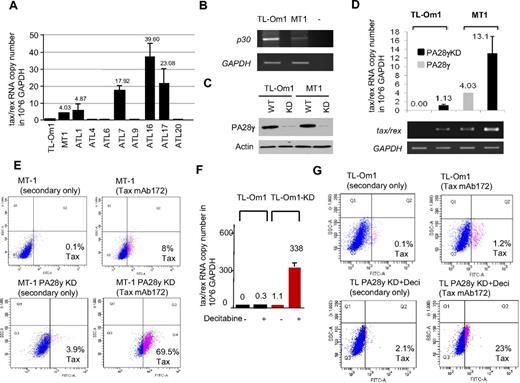
We next wanted to validate our results in human T cells infected with HTLV-1. Because PA28γ is required for p30 to control virus replication, we hypothesized that reducing PA28γ expression may be sufficient to reactivate virus expression in ATLL cells latently infected with HTLV-1. To this end, we used HTLV-1–infected T-cell lines derived from ATLL patients. These ATLL-derived lines harbor latent integrated provirus and express no detectable levels of viral RNA (TL-Om1) or low levels of tax/rex RNA (MT-1; Figure 6A). Interestingly, p30 RNA expression was inversely correlated with tax/rex expression in these cells (Figure 6B). This observation is reminiscent of ATLL patients and studies that have shown frequent methylation of the viral 5′ LTR in vivo. TL-Om1 has also been shown to harbor a methylated 5′ LTR. In agreement, analyses from 8 primary ATLL uncultured samples for tax/rex expression showed that one-half of patients have no detectable expression (similar to TL-Om1) and one-half have detectable tax/rex expression (similar to MT-1). MT-1 and TL-Om1 cells were infected with lentiviral vectors expressing PA28γ shRNA and selected with puromycin. Knock-down (KD) of PA28γ from MT-1 (KD) and TL-Om1 (KD) cell lines compared with their parental lines was confirmed by Western blot (Figure 6C). Interestingly, real time quantitative Taqman RT-PCR demonstrated an increase of the tax/rex RNA copy number (per million GAPDH) from 4 to 13 in MT-1 and MT-1 (KD) cells (Figure 6D). The increase of tax/rex RNA in TL-Om1 (KD) was small, suggesting that targeting PA28γ alone may not be sufficient in ATLL cells harboring a 5′ methylated LTR. To confirm the biologic significance of our results in MT1 (KD), we next investigated if viral antigens were re-expressed and detected at the protein level using intracellular staining and FACS analyses for Tax. Our results showed a significant increase in Tax expression in MT-1 (KD) after inhibition of PA28γ expression, from 8% to 69.5% of Tax-positive cells, compared with MT-1 parental cells analyzed in the same conditions (Figure 6E). These findings are highly significant because previous studies highlighted the fact that Tax is very immunogenic and the dominant viral protein targeted by the CTL response in HTLV-1–infected patients.25 Our observations suggest that targeting PA28γ may be effective in breaking latency in some ATLL patients. Expression of Tax protein was undetectable in TL-Om1 (KD) with a signal similar to background levels, (Figure 6G). We hypothesized that methylation could exert a block and tested the effects of demethylating agents. Treatment of parental TL-Om1 with decitabine had limited effect on virus re-expression, which remained below the threshold of Tax protein detection as shown by quantification of tax/rex RNA (Figure 6F). However, treatment with decitabine had a synergistic effect with the inhibition of PA28γ and demonstrated re-expression of tax/rex RNA in TL-Om1 (KD) cells (Figure 6F-G). We did not observe any change in HBZ RNA expression comparing MT-1 and MT-1 PA28γ KD, or TL-Om1 KD and TL-Om1 KD treated with decitabine (data not shown). These observations suggest that the increase in tax/rex mRNA expression is not associated with changes in transcription from the 3′ LTR. Altogether, our results suggest that in ATLL patients with a 5′ methylated LTR, combination therapy targeting both DNA demethylation and PA28γ inhibition may be a very effective approach to breaking latency. Future studies aimed at designing small inhibitors specific to PA28γ are warranted. Reactivation of virus expression from latently infected ATLL cells will offer a unique opportunity to unmask and, in combination with therapeutic vaccines, eradicate HTLV-1–infected cells from the host.
Reactivation of viral gene expression from HTLV-1 latently infected ATLL cells. (A) Taqman quantification of tax/rex RNA copy number per million GAPDH in MT-1, TL-Om1, and 8 additional uncultured ATLL samples. Standard deviations were calculated from 4 independent experiments. (B) RT-PCR detection of p30 RNA expression in TL-Om1 and MT-1 cells using LTR2 and TF4 for 43 cycles. (C) Western blot analyses showing knockdown (KD) of PA28γ expression in MT-1KD and TL-Om1KD cell lines. (D) Taqman quantification of tax/rex RNA copy number per million GAPDH in MT-1, MT-1KD, TL-Om1, and TL-Om1KD cells. Standard deviations were calculated from 2 independent experiments. Agarose gel amplification is shown underneath. (E) Intracellular staining for detection of Tax expression by FACS in MT-1 parental cell line and MT-1KD cells using Tax monoclonal antibody Tab172. (F) Taqman quantification of tax/rex RNA copy number per million GAPDH in TL-Om1 and TL-Om1KD treated with decitabine (1μM) for 72 hours. Standard deviations were calculated from 2 independent experiments. (G) Intracellular staining for detection of Tax expression by FACS in TL-Om1 parental cell line and in TL-Om1KD cells treated with decitabine using Tax monoclonal antibody Tab172.
Reactivation of viral gene expression from HTLV-1 latently infected ATLL cells. (A) Taqman quantification of tax/rex RNA copy number per million GAPDH in MT-1, TL-Om1, and 8 additional uncultured ATLL samples. Standard deviations were calculated from 4 independent experiments. (B) RT-PCR detection of p30 RNA expression in TL-Om1 and MT-1 cells using LTR2 and TF4 for 43 cycles. (C) Western blot analyses showing knockdown (KD) of PA28γ expression in MT-1KD and TL-Om1KD cell lines. (D) Taqman quantification of tax/rex RNA copy number per million GAPDH in MT-1, MT-1KD, TL-Om1, and TL-Om1KD cells. Standard deviations were calculated from 2 independent experiments. Agarose gel amplification is shown underneath. (E) Intracellular staining for detection of Tax expression by FACS in MT-1 parental cell line and MT-1KD cells using Tax monoclonal antibody Tab172. (F) Taqman quantification of tax/rex RNA copy number per million GAPDH in TL-Om1 and TL-Om1KD treated with decitabine (1μM) for 72 hours. Standard deviations were calculated from 2 independent experiments. (G) Intracellular staining for detection of Tax expression by FACS in TL-Om1 parental cell line and in TL-Om1KD cells treated with decitabine using Tax monoclonal antibody Tab172.
Discussion
Human RNA and DNA tumor viruses have evolved multiple strategies to evade immune defenses and persist in their host by establishing a latent infection and viral reservoirs that evade immune recognition.26 Proliferation is a necessity for pre-tumor cells to accumulate genetic alterations and to acquire a transformed phenotype. However, cell activation and division is often associated with transient expression of viral antigens and the risk of being recognized and eliminated by host immune defenses. Therefore, latent viruses need to cope with sufficient expression of tumor-associated viral factors while keeping a tight suppression of immunogenic viral proteins. In addition to tumor viruses, the human immunodeficiency virus (HIV) also establishes a lifelong infection. Although current antiretroviral therapy (ART) efficiently controls HIV-1 replication, it fails to eliminate the virus.27 Even after years of successful ART, HIV-1 can conceal itself in a latent state in long-lived CD4 (+) memory T cells. From this latent reservoir, HIV-1 reemerges if ART treatment is interrupted.27 Attempts to eradicate viral reservoirs have repeatedly yielded disappointing results. Dendritic cell-based vaccines are promising as they induce a broad and long-lasting immunity. Unfortunately, it may not prevent viruses from establishing long-term latency, and as a result, may not reduce the risk of cellular transformation. Similarly, the use of live-attenuated vaccines limits but does not eliminate long-term latency. For these reasons, a better understanding of viral latency and the cellular genes involved in this process is crucial for the development of new therapeutic approaches aimed at the elimination of latent viral reservoirs. Conceivably, establishment of latency involves mainly viral decisions and viral genes, whereas the maintenance of viral latency is mainly controlled by cellular genes or events.
The discovery of p30-mediated repression of HTLV-1 replication suggests that retroviruses have evolved specific viral factors to hide in the host. We previously found that the HTLV-1 p30 protein modulates viral expression and helps the virus to remain concealed and avoid elimination by the host immune system. We have now identified an intriguing new molecular mechanism used by the HTLV-1 protein p30 to suppress viral gene expression and to inhibit virus replication. Our results identify for the first time the proteasome activator PA28γ as a cellular factor involved in viral mRNA nuclear retention and acting as corepressor of HTLV-1 replication. Consequently, tax/rex mRNA nuclear export is interrupted and viral tax/rex mRNA accumulates in the nucleus. Other studies have suggested that p30 interactions with PA28γ may also affect ATM functions and increase cell survival.28 This may be important because p30 has also been shown to affect cell cycle progression and to prevent homologous DNA repair by targeting the MRN complex.29,30
Mechanistically, we found that p30 specifically interacts with PA28γ and recruits the latter onto the tax/rex viral mRNA. Mapping experiments uncovered a small distinct region of PA28γ responsible for interactions with p30. Mutagenesis experiments were performed to generate a PA28γ mutant no longer able to interact with p30. This mutant was used to demonstrate that interactions between these 2 proteins are required for p30 repressive effects on HTLV-1 replication. Notably, using distinct HTLV-1 molecular clones and PA28γ knockdown cells, we showed that p30-mediated repression of HTLV-1 replication occurs in a natural context with physiologic expression of p30. Furthermore, the effect is specific because a p30-mutated molecular clone replicated in a similar fashion in PA28γ knockdown or parental cells. Overall, our studies identify the cellular factor PA28γ as a critical corepressor of HTLV-1 replication and virus silencing, a key step in the establishment of viral latency. Our findings suggest that reactivation of latent HTLV-1–infected cells may be a useful strategy, a concept we proposed almost 10 years ago. Recently, the use of valproate with AZT confirmed that reactivation and eradication of infected cells in vivo is possible.31 Whether PA28γ may also be involved in the control of other virus-induced latency is an exciting possibility and warrants additional studies. Our findings that HTLV-2 p28 was unable to interact with PA28γ and could not suppress HTLV-2 replication are intriguing because HTLV-2 has not been associated with diseases in humans. It is possible that p30 and PA28γ interactions are required for establishment of a specific reservoir playing a role in HTLV-1 pathogenesis. Because proteasome functions of PA28γ are required for suppression of viral replication, this suggests that the p30/PA28γ complex may lead to the degradation of specific cellular factor(s) involved in viral mRNA export. Identification of such factor(s) is critically important and may shed light on a new mode of repression used in cells to control mRNA nuclear export. Along these lines, previous studies have shown that PA28γ can facilitate, in a substrate selective manner, the degradation of specific proteins by proteasomes.32
Disrupting PA28γ interactions with p30 or the use of small inhibitors designed to specifically block the proteasome activity of PA28γ may be sufficient to reactivate virus expression and eradicate HTLV-1 latently infected cells in vivo in a subset of ATLL patients expressing tax/rex mRNA as shown by our data using MT-1 ATLL cells. We found approximately one-half of ATLL patients expressing the tax/rex mRNA in vivo. These results are consistent with previous findings that the 5′LTR is frequently hypermethylated. Our results demonstrate that in ATLL TL-Om1 cells, which have a methylated LTR, no tax/rex expression is detectable. Surprisingly, treatment with demethylating agents or knocking down PA28γ expression had little effect on tax/rex mRNA reactivation, which remained below the threshold of protein detection. However, combination treatment targeting both methylation and PA28γ had a radical effect and strongly reactivated HTLV-1 viral antigen expression in TL-Om1 knocked down for PA28γ expression. Our data support the feasibility of breaking latency in HTLV-1–transformed ATLL cells and the possibilities raised by this concept could lead to a completely new approach to multiple drug treatment of HTLV-1 retroviral diseases.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Brandi Miller for editorial assistance and Dr G. Pratt and Dr M. Rechsteiner (University of Utah) for their assistance with the in vitro proteasome assays.
This work was supported by National Institute of Allergy and Infectious Diseases (NIAID) grant AI058944 to C.N.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID or the National Institutes of Health.
National Institutes of Health
Authorship
Contribution: N.L.K., J.M.T., M.B., X.T.B., S.P.S., and M.D. performed research experiments; and C.N. designed the study, interpreted the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christophe Nicot, University of Kansas Medical Center, Dept of Pathology and Laboratory Medicine, Center for Viral Oncology, 3901 Rainbow Blvd, Kansas City, KS 66160; e-mail: cnicot@kumc.edu.