Key Points
Mutations of key transcription factor in myeloid malignancies.
Abstract
Congenital neutropenia is a group of genetic disorders that involve chronic neutropenia and susceptibility to infections. These neutropenias may be isolated or associated with immunologic defects or extra-hematopoietic manifestations. Complications may occur as infectious diseases, but also less frequently as myelodysplastic syndrome (MDS) or acute myeloid leukemia (AML). Recently, the transcription factor GATA2 has been identified as a new predisposing gene for familial AML/MDS. In the present study, we describe the initial identification by exome sequencing of a GATA2 R396Q mutation in a family with a history of chronic mild neutropenia evolving to AML and/or MDS. The subsequent analysis of the French Severe Chronic Neutropenia Registry allowed the identification of 6 additional pedigrees and 10 patients with 6 different and not previously reported GATA2 mutations (R204X, E224X, R330X, A372T, M388V, and a complete deletion of the GATA2 locus). The frequent evolution to MDS and AML in these patients reveals the importance of screening GATA2 in chronic neutropenia associated with monocytopenia because of the frequent hematopoietic transformation, variable clinical expression at onset, and the need for aggressive therapy in patients with poor clinical outcome.
Introduction
Congenital neutropenia is a group of genetic disorders that involve chronic neutropenia and susceptibility to infections. Mutations have been described previously in congenital neutropenia that is isolated (eg, mutations of ELANE), associated with immunologic defects (eg, mutations of CXCR4 or WAS genes), or involves extra-hematopoietic manifestations, the most frequent being HAX1 (Kostmann disease), SDBS (Shwachman-Bodian-Diamond Syndrome), or G6PC3.1 Complications may occur in congenital neutropenia, essentially infectious diseases but also less frequently myelodysplastic syndrome (MDS) or acute myeloid leukemia (AML). The rate of evolution of patients with severe congenital neutropenia is 10.8% at 20 years increasing to 18.8% in patients with SBDS mutations.2
MDS is a clonal hematopoietic disorder resulting in ineffective hematopoiesis and frequently evolving to AML. Most frequently, MDS and AML are sporadic and affect primarily elderly patients with a median age more than 65 years. In contrast, genetic susceptibility to MDS and/or AML is infrequent. RUNX1 and CEBPA, 2 oncogenes encoding transcription factors, have been linked to familial AML and MDS.3 Germline heterozygous mutations of RUNX1 occur mostly in the Rel homology domain, leading to the loss of its DNA-binding capacity3 and being associated with a familial platelet disorder and an autosomal-dominant transmission.4 CCAAT enhancer binding protein α (CEBPA) is mutated in familial AML with an autosomal-dominant transmission and a high penetrance.5 In addition to RUNX1 and CEBPA, mutations in the telomerase RNA (TERC) or reverse transcriptase (TERT) subunits have also been identified.6
Recently, GATA2 has been identified as a new predisposing gene for familial AML/MDS.7-11 GATA2 is a member of the GATA transcription factor family characterized by a 2 zinc finger domains binding to the DNA sequence WGATAR and a transactivation domain. Heterozygous point mutations located in the second zinc finger of GATA2 have been identified initially in 4 families with MDS and AML.7 Point mutations have been also identified in diseases that overlap with familial AML/MDS, such as Emberger8 and MonoMAC syndromes.12,13 Emberger syndrome is an autosomal-dominant primary lymphedema with a predisposition to AML.14,15 The MonoMAC syndrome is characterized by a severe deficiency of monocytes, B-lymphocytes, and natural killer (NK) cells, along with nontuberculous mycobacterial, fungal, and viral infections and an autosomal-dominant transmission in most cases.10
In the present study, we describe the initial identification by exome sequencing of GATA2 mutation in a mother and her 3 children associated with a prior history of chronic mild neutropenia evolving to AML and/or MDS. Subsequent analysis of the French Severe Chronic Neutropenia Registry allowed the identification of 10 additional patients with GATA2 mutations in 6 pedigrees with unexplained mild neutropenia in at least 1 patient of the pedigree.1,2,16
The frequent evolution to MDS and AML of patients with GATA2 mutations reveals the importance of screening for this mutation in chronic neutropenia, especially when associated with monocytopenia because of frequent hematopoietic transformation, variable clinical expression at onset, and the need of aggressive strategy therapy in patients with poor clinical outcome.
Methods
Patients
Patient pedigree 1 samples were obtained after written informed consent in accordance with the Declaration of Helsinki and were stored at the HIMIP (Hématologie-Inserm Midi-Pyrénées) collection (declared to the Ministry of Higher Education and Research as DC 2008-307 collection 1). Clinical and biologic annotations of the samples have been declared to the Comité National Informatique et Libertés.
French Severe Chronic Neutropenia Registry
Patients were registered in the French Severe Chronic Neutropenia Registry, which was established in 19931 to collect data from 35 pediatric hematology-oncology clinical units. The inclusions criteria of the registry are: neutrophil count lower than 0.5 G/L observed at least 3 times in a period of 3 months or a neutrophil count lower than 1 G/L observed at least 3 times in a period of 3 months associated with infection. Systematic recording of infectious episodes was obtained from the patients' written medical history. Severe infections were defined as those that would be life-threatening without appropriate antibiotic or antifungal therapy. Minor infections were defined as those for which patients did not seek medical attention (eg, stomatologic, ear, nose, and throat infections). Warts were considered when they numbered ≥ 10 or when they presented as genital condylomata acuminata. The patients or their parents gave their written informed consent for genetic testing. We selected for GATA2 by screening 14 probands from a series of 544 patients included in the registry. The selection criteria were the absence of mutations for the major genes involved in congenital neutropenia (ELANE, CXCR4, SBDS, HAX1, and G6PC3; n = 288 patients) and the presence of clinical features associated with GATA2 mutations: leg lymphedema, warts, and evolution to MDS/AML. The median age at the last follow-up was 18 years (range, 7-56).
Karyotype
Karyotypes were established from bone marrow (BM) aspiration using standard procedures and described according to the International System for Human Cytogenetic Nomenclature.
CGH analysis
For comparative genomic hybridization (CGH) analysis, genomic DNA samples were analyzed for copy number changes using Affymetrix Genome-Wide Human SNP 6.0 arrays (for pedigree 1) or CytoScan HD arrays (for other pedigrees when material was available). Sample preparations and hybridizations were performed using Genome-wide Human SNP Nsp/Sty and CytoScan assay kits (Affymetrix) according to the manufacturer's protocol. Analysis of copy number state was done using BRLMM-P-Plus algorithm with regional GC correction, embedded in Genotyping Console Version 2.0 and Chromosome Analysis Suite software (Affymetrix).
Exome sequencing
A total of 3 μg of DNA extracted from the BM sample of patient number 6225 (blast infiltration, 30%) was fragmented by sonication to 150-200 bp and ligated to an oligonucleotide adaptor. Fragments were purified and amplified by PCR for 6 cycles. Five hundred nanograms of this PCR product were hybridized to the SureSelect Human All Exon Kits Version 2 (Agilent Technologies), covering 44 Mb of the human exome during 24 hours. After washing, the eluate was amplified by PCR for 12 cycles. The sequencing of the exome was performed by IntegraGen using a HiSeq 2000 (Illumina). Base calling was performed using Illumina RTA Version 1.8 using default settings. Sequencing data were analyzed with Illumina CASAVA1.7 software and the human genome reference sequence hg19. Average depth was 124×. Single-nucleotide variants (SNVs) were filtered using COSMIC v53 and dbSNP135 databases for nonsynonymous variants with a reading call higher than 5, leading to the identification of 8 SNVs: EPHB6 T483P, ERN V458G, GATA2 R396Q, IMPG2 P1161L, KDM5C V858G, MAST4 K2422E, NEK11 D87Y, and TTN P4737R. Five were confirmed by classic Sanger sequencing: EPHB6 T483P, GATA2 R396Q, IMPG2 P1161L, NEK11 D87Y, and TTN P4737R (primer sequences are provided in supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
GATA2 mutation screening
GATA2 mutations in exons 2-6 were searched for by PCR and Sanger sequencing (primer sequences are listed in supplemental Table 1). Sequences were analyzed with Sequencher (Genescode). Mutations are numbered as recommended by the Human Genome Variation Society (http://www.hgvs.org/) using the reference sequence NM_032638.4.
Chromosome 3 microsatellite genotyping
The familial segregation of the GATA2 R396Q mutation (GATA2 gene location according to hg19, chr3:128.2 Mb) in pedigree 1 was investigated using 4 highly polymorphic short tandem repeats (STRs), 2 of which were centromeric to GATA2 (D3S3606 and D3S3607, located at chr3:127.2 Mb and chr3:127.3 Mb, respectively) and 2 of which were telomeric to GATA2 (D3S1587 and D3S1292 located at chr3:130.8 Mb and chr3:131.6 Mb, respectively). STRs were amplified by PCR (primer sequences are provided in supplemental Table 1) and analyzed by GeneScan analysis performed on an Applied Biosystems 3130XL.
Statistical methods
Stata Version 10 software was used for all statistical analyses. The median, lower, and upper interquartile values depict the distribution of quantitative variables. For survival analyses, end points were death and the first episode of cancer. The period taken into account was the time interval from birth to the first date when the event was observed or when no event occurred to the last examination. The Kaplan-Meier method was used to estimate survival rates. The cutoff date was July 1, 2012.
Results
Clinical presentations of pedigree 1
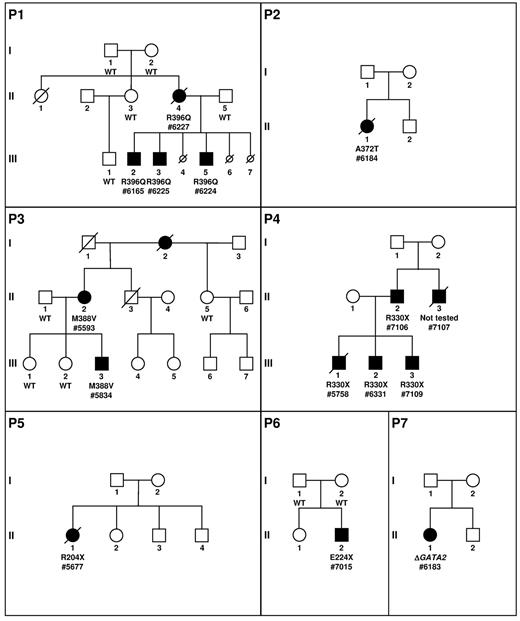
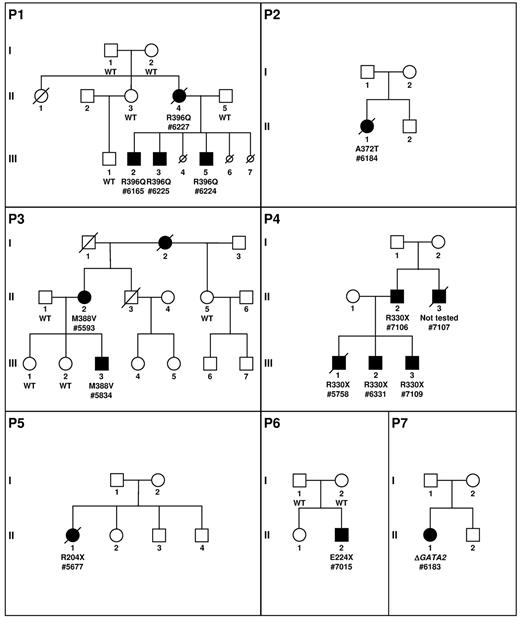
We investigated a family in which the mother (patient number 6227; P1, II, 4) developed at 35 years of age an AML M2 associated with myelodysplasia-related changes. Karyotype was normal. She had a history of recurrent gingivitis and skin infection consistent with chronic neutropenia (12 years before AML) and monocytopenia. She presented with aspergillosis during induction therapy and received several lines of chemotherapy because of a refractory disease. She died from CMV pneumonia without achieving complete remission.
The patient had 3 sons and 2 miscarriages. Three years later, at age 14, her second child (patient number 6225; P1, III, 3) developed an AML M2 associated with myelodysplasia-related changes with a similar cytologic presentation to his mother. The karyotype identified a trisomy 11 and a partial deletion of chromosome 7. He had a previous history of recurrent infections and mild neutropenia. In first complete remission, he received a matched unrelated donor hematopoietic stem cell transplantation (HSCT) and is still in remission 18 months after the allograft. His elder brother (patient number 6165; P1, III, 2), 16 years of age, also had a history of chronic neutropenia with a chronic EBV replication in the gut and a severe respiratory distress syndrome requiring supplemental oxygen at age 14 linked to pulmonary bronchiectasis. Examination of the blood and the BM revealed a refractory cytopenia with multilineage dysplasia. The karyotype was normal. A global immunologic defect was identified with a very low number of CD14+ monocytes, CD19+ B-cell lymphocytes, and CD3−/CD56+ NK cells, along with a normal CD3+ T-cell lymphocyte count (supplemental Table 2). He underwent HSCT with a matched unrelated donor in April 2012. The youngest brother (patient number 6224; P1, III, 5), 5 years of age, presented with recurrent fever and chronic EBV replication in the blood. Blood count and BM examination were normal. Karyotype identified a complete monosomy 7 leading to the diagnosis of unclassifiable MDS (MDS-U). Matched unrelated BM transplantation was performed and was complicated by a grade 4 cutaneous GVHD requiring intensive immunosuppressive therapy. Fourteen months after the transplantation, no severe infection was noted and immunologic recovery was obtained.
Identification of a germline GATA2 R396Q mutation in pedigree 1
Because a familial MDS/AML was suspected in pedigree 1, we looked for a putative germline mutation. RUNX1 and CEBPA were not mutated in any member of this family (data not shown). No recurrent deletion, amplification, or uniparental disomy was detected by CGH (data not shown). Therefore, we performed exome sequencing on the AML diagnosis sample of the second son (patient number 6225). Five SNVs were identified further and confirmed by classic Sanger sequencing: EPHB6 T483P, GATA2 R396Q, IMPG2 P1161L, NEK11 D87Y, and TTN P4737R. We examined the other members of the family (mother, father, the 2 other sons, maternal grandfather, maternal grandmother, and maternal cousins) for the presence of these variations. Two variations, GATA2 R396Q (c.1187G A; supplemental Figure 1) and NEK11 D87Y, both genes located on the chromosome 3 that are separated by 2.7 Mb, were identified in the mother and in the 3 children's samples but were absent in the father's sample. The GATA2 R396Q variation was not identified in the maternal grandparents' DNA, but the NEK11 D87Y variation was detected in the maternal grandfather's DNA sample. Because the maternal grandfather had no clinical or biologic phenotypes, we focused our analysis on the GATA2 R396Q mutation. We investigated its origin using 4 highly polymorphic short tandem repeats, 2 centromeric to GATA2 (D3S3606 and D3S3607) and 2 telomeric to GATA2 (D3S1587 and D3S1292). Genotyping STR analysis demonstrated that the chromosome 3 allele bearing the GATA2 R396Q mutation came from the maternal grandfather, although the mutation was not detected in his DNA sample (supplemental Figure 2), suggesting a de novo event in patient II-4 (patient number 6227).
GATA2 mutations are identified in chronic neutropenia
Because the clinical presentation of pedigree 1 was initially chronic neutropenia, we investigated the presence of mutations of GATA2 in the French Severe Chronic Neutropenia Registry. Fourteen unrelated proband cases were selected for the presence of a neutropenia, a WHIM-like syndrome with possible evolution to MDS/AML, and their negativity for mutations of the major genes involved in congenital neutropenia (ELANE, CXCR4, SBDS, and G6PC3). We identified 6 mutations of GATA2 among these 14 cases, 5 point mutations and a large deletion of 61 kb completely overlapping the GATA2 locus. In 2 of these 6 pedigrees (P3 and P4, Figure 1), we identified a GATA2 mutation in 4 affected relatives, 1 in P3 and 3 in P4. Overall, we detected a GATA2 mutation in 14 white patients.
Pedigree description. P1 to P7 describes the pedigrees. Circles represent females and squares males. Parents are connected by a single horizontal line, and vertical lines indicate their offspring. Offspring are connected by a horizontal line. Siblings are placed from left to right according to birth order and are labeled with Arabic numerals. Each generation is indicated by a Roman numeral. Circles and squares are black when neutropenias have been formally identified. If a patient is dead, a diagonal line is placed over the circle or square. Smaller circles with a diagonal line indicate miscarriage (P1, III, 4; P1, III, 6) or termination because of AML induction treatment (P1, III, 7). Patients tested are indicated by the result of the GATA2 screening and a number (patient number 7107 was not analyzed). The absence (wild-type, WT) or presence of a GATA2 mutation (protein variation or complete deletion indicated by Δ) is indicated below the symbol of the patient tested.
Pedigree description. P1 to P7 describes the pedigrees. Circles represent females and squares males. Parents are connected by a single horizontal line, and vertical lines indicate their offspring. Offspring are connected by a horizontal line. Siblings are placed from left to right according to birth order and are labeled with Arabic numerals. Each generation is indicated by a Roman numeral. Circles and squares are black when neutropenias have been formally identified. If a patient is dead, a diagonal line is placed over the circle or square. Smaller circles with a diagonal line indicate miscarriage (P1, III, 4; P1, III, 6) or termination because of AML induction treatment (P1, III, 7). Patients tested are indicated by the result of the GATA2 screening and a number (patient number 7107 was not analyzed). The absence (wild-type, WT) or presence of a GATA2 mutation (protein variation or complete deletion indicated by Δ) is indicated below the symbol of the patient tested.
Patient number 6184 (P2, II, 1) was diagnosed at the age of 13 with an AML M5. The karyotype identified a translocation involving MLL along with a monosomy 7. She underwent treatment with intensive chemotherapy that was complicated by pulmonary aspergillosis. Her brother and parents are healthy. A complete remission was obtained, but she developed a progressive loss of neutrophils, monocytes, B cells, and NK cells associated with numerous infections (Table 1 and supplemental Table 2). Genetic evaluation finally identified a heterozygous missense GATA2 A372T mutation (c.1114G > A, supplemental Figure 1). The patient died 5 years later from an H1N1 influenza infection in complete remission.
Patient number 5593 (P3, II, 2) developed chronic neutropenia at the age of 30 associated with numerous HPV-induced warts since age of 14 years. The BM smear did not identify myelokathexis features and the case was considered to be a WHIM-like syndrome. She suffered from a chronic genital papillomavirus infection evolving to carcinoma. She had additional infections such as pulmonary aspergillosis, recurrent oral herpes, and onychomycosis infections. A deficit of monocytes, B cells, and NK cells was detected, the T-cell count remained normal (Table 1 and supplemental Table 2). We identified a heterozygous missense GATA2 M388V mutation (c.1162A > G, supplemental Figure 1). She had 3 children, 1 of whom developed MDS detected in the evaluation of asthenia at the age of 12 (patient number 5834; P3, III, 3). He was treated by HSCT and is still in complete remission 2.5 years after the transplantation.
Patient number 5758 (P4, III, 1) presented with numerous and extensive warts associated with a mild neutropenia since the age of 6, a profound monocytopenia, and deficiencies of B cells and NK cells (Table 1 and supplemental Table 2). He developed, at the age of 18, AML with myelodysplastic features associated with a der(1;7)(q10;p10) leading to a monosomy 7q and a trisomy 1q. A heterozygous nonsense GATA2 R330X mutation (c.988C > T, supplemental Figure 1) was identified. He presented with an Aspergillus infection during induction therapy and died from severe respiratory distress. His younger brother (patient number 6331; P4, III, 2) presented recurrent otitis during infancy and had a long period of extensive warts since the age of 12. Blood and medullar tests revealed mild neutropenia; monocytopenia; B-, NK-, and dendritic-cell deficiencies; and an hypoplastic BM with a karyotype harboring the same chromosomal alteration observed in the BM of his brother, a der(1;7)(q10;p10) leading to a partial monosomy 7q. At 20 years of age, he underwent HSCT with an unrelated donor. He is still in hematologic and cytogenetic remission 2 years after HSCT. His younger brother (patient number 7109; P4, III, 3) presented at the age of 15 with severe meningococcal infection associated with mild hematologic abnormalities and an identical GATA2 mutation. Blood and medullar tests revealed a mild neutropenia and thrombocytopenia, B-cell deficiencies, and a very slightly hypoplastic BM with a normal karyotype. The father (patient number 7106; P4, II, 2) has the same mutation. Blood and medullar tests were normal except for a mild B-cell deficiency. The paternal uncle (patient number 7107; P4, II, 3), although not tested, has a presentation evocating a GATA2 mutation with recurrent warts associated with mild neutropenia and monocytopenia. He died suddenly at the age of 36 from aortic dissection.
Patient number 5677 (P5, II, 1) presented with WHIM-like syndrome with HPV-induced warts, clinical features of extended sarcoidosis, neutropenia, lymphopenia, monocytopenia, and a severe defect in NK cells.17 She developed a disseminated Mycobacterium avium complex infection and later an AML M4 subtype. She died from a refractory disease after several lines of chemotherapy. We identified a heterozygous GATA2 R204X mutation (c.610C > T, supplemental Figure 1).
Patient number 7015 (P6, II, 2), born from healthy parents, presented with several warts in the extremities and on the forehead at the age of 4. A routine blood test performed at 11 years of age revealed a mild chronic neutropenia, monocytopenia, and mild lymphopenia with a profound B-cell defect (Table 1 and supplemental Table 2). We identified a heterozygous nonsense GATA2 E224X mutation (c.670G > T, supplemental Figure 1) that was not detected in the parents' samples. Initial treatment based on G-CSF and corticosteroids showed a moderate efficacy on neutropenia.
Patient number 6183 (P7, II, 1) is a young girl with an initial presentation of neutropenia evolving to MDS associated with the loss of chromosome 7 (Table 1 and supplemental Table 2). She had a large monoallelic deletion of 61 kb (Chr3:128.17-128.23 Mb) completely encompassing GATA2 and 2 other genes, DNAJB8 and LOC90246 (supplemental Figure 1). She was allografted and is still alive 1 year later.
Preleukemic hematologic and immunologic parameters
Before the evolution to MDS or AML, a median of 4 blood counts were available per patient. In all cases, hematologic parameters fluctuated with time, except for monocytes, which remained low. The median neutrophil count was 1.5 G/L (25th percentile: 1.2; 75th percentile: 2.0), although the count was mildly diminished, all patients experienced at least once a neutropenia between 0.5 and 1.2 G/L. The monocyte count was very low, with a median value of 0.05 G/L (25th percentile: 0.03; 75th percentile: 0.17). The lymphocyte count was also low, with a median at 1.1 G/L (25th percentile: 1.0; 75th percentile: 1.7); with B cells and NK cells were also low. The hemoglobin and platelet counts were at the lower range of normal values with a median of 12.5 g/dL (25th percentile: 11.2; 75th percentile 13.4) and 152 G/L (25th percentile: 125; 75th percentile: 180), respectively. BM smears were available at baseline for 4 patients. The differential counts were within the normal range without evident myeloid cytologic abnormalities. The immunologic standard evaluation, even though it was not systemically performed, revealed mild abnormalities (supplemental Table 2). Immunoglobulin levels were almost in the normal range. In the 2 cases studied (patient number 6165 and 6184), the vaccine antigen response was normal (supplemental Table 2).
Associated morbidities
Overall, the main clinical features identified in these patients are the presence of warts at the initial diagnosis, frequent infections, and evolution to MDS and/or AML. Two patients (patient numbers 5677 and 6184) had infection profiles compatible with MonoMAC syndrome. Recurrent pneumonia was present in 5 patients and severe and chronic warts and/or genital HPV was observed in 6 patients. The induction therapy of AML was complicated by lung aspergillosis in 2 patients of 4 who have received this therapeutic approach, and 1 additional patient (patient number 5593) suffered from lung aspergillosis along with genital cancer. Among the 3 older patients, 2 had presented severe vascular complications, 1 with a lethal aortic dissection at the age of 36 (patient number 7107) and 1 cardiac infarct at the age of 50 in a woman without risk factors (patient number 5593). One patient (patient number 6183) presented with an urethral malposition that was corrected at the age of 2, and 1 patient (patient number 6225) presented with a thigh fibroma at the age of 3. In our series, none of the patients bearing GATA2 mutations exhibit lymphedema or deafness.
Clinical evolution
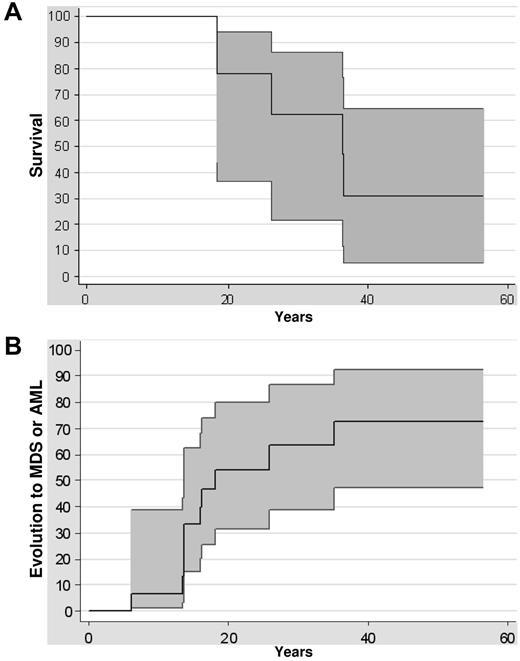
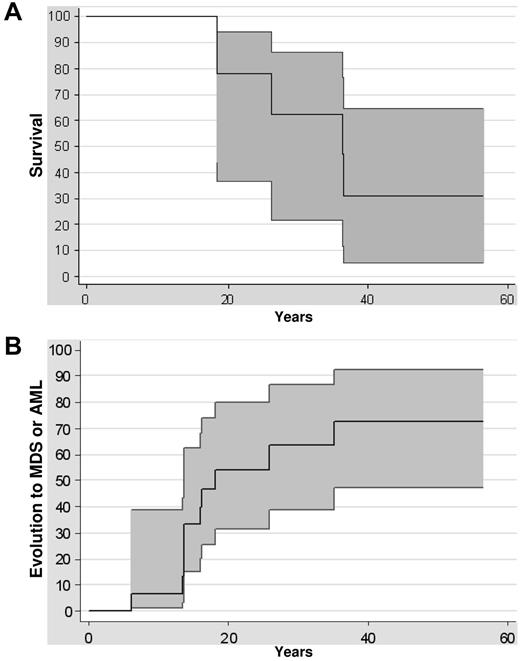
Survival rates were 77% (at 20 years) and 62% (at 30 years) in this series of 14 patients with GATA2 mutations (Figure 2A). Five deaths were recorded, 3 as a direct consequence of the leukemic evolution (patient numbers 6227, 5758, and 5677), 1 related to a flu-mediated pneumonia in a patient in complete remission of AML with immunodeficiency (patient number 6184), and 1 in a patient who died from an aortic dissection at the age of 36 (patient number 7107). The risk of malignant transformation was extremely high, because 6 of 14 patients with GATA2 mutations developed a refractory cytopenia with multilineage dysplasia and 4 patients developed AML (Figure 2B). The median age at the diagnosis of MDS or AML was 15 years (range, 7-35). The risk of transformation at the age of 10, 20, or 30 was, respectively, 6%, 54%, and 63% (Figure 2B). The outcome once the MDS or the AML occurred was extremely poor: none of the patients treated only by chemotherapy was alive in contrast to the 6 patients receiving HSCT (Table 1).
Clinical evolution. (A) Kaplan-Meier survival curve of GATA2-mutated patients. Confidence intervals of 95% are indicated. (B) Evolution of GATA2-mutated patients to MDS or AML from birth to evolution to MDS or AML or last follow-up. Confidence intervals of 95% are indicated.
Clinical evolution. (A) Kaplan-Meier survival curve of GATA2-mutated patients. Confidence intervals of 95% are indicated. (B) Evolution of GATA2-mutated patients to MDS or AML from birth to evolution to MDS or AML or last follow-up. Confidence intervals of 95% are indicated.
Discussion
We identified 7 different GATA2 mutations in 14 patients coming from 7 pedigrees (Figure 1). The molecular consequences of the mutations identified here can be classified in 2 distinct groups, the first with mutations located in or close to the second GATA2 zinc finger (A372T mutation in P2 located between the third [C370] and fourth [C373] cysteines of the zinc finger, M388V in P3 and R396Q in P1) and the second with the complete loss of 1 allele of GATA2 (P7) or creation of a stop codon leading to the loss of the second zinc finger (R330X mutation in P4 located in the first zinc finger, R204X [P5] and E224X [P6] mutations leading to the loss of the 2 zinc fingers). These nonsense mutations are likely be sensitive to nonsense-mediated decay and would be null alleles similar in effect to whole gene deletions. Only the mutation R396Q in P1 has been described previously in 1 case of MonoMAC syndrome.18
Mutations of GATA2 described previously7-10,12,13,19,20 are highly heterogeneous (supplemental Table 3), ranging from the creation of a stop codon (R337X), a frameshift mutation scattered over the whole coding frame (R78, G81, L105, A194, G200, D259, N317, L332 and A341), deletions (ranging from large deletions encompassing the whole GATA2 locus to smaller deletions such as codons 1-290, codons 340-381, codons 341-346, and codons 362-365 or T355), and point mutations (located close to the second zinc finger domain with the exception of P254L: P304H, C319SC, T344M, T354M, L359V, R361L, R362Q, N371K, C373R, R396W, R396Q, and R398W). The majority of GATA2 mutations have been identified in MDS/AML, Emberger, or MonoMAC syndrome patients.7,8,12,13,20 The L359V mutation and the deletion of codons 341-346 have been exclusively identified in myeloid transformation of chronic myeloid leukemias and are acquired.19 Only 3 of these mutations (L359V, T354M, and M355del) have been functionally tested by gel-shift or luciferase assays.7,19 The L359V has an opposite effect to the T354M and M355del mutations, with a gain of function effect for the former and a loss of function for the latter. The spectrum of GATA2 mutations in MDS/AML, Emberger, or MonoMAC syndrome patients is large.19 In our cases and in other cases reported with germline GATA2 mutations, no clear correlation between genotype and phenotype can be identified, suggesting that these clinical presentations are a spectrum of the same disease.7-9,12,13
The clinical evolution in MDS or AML of patients with germline GATA2 mutations is frequently associated with a partial or complete deletion of chromosome 7: 6 of 11 patients in our study (2 with the R330X mutation, 1 with the A372T mutation, 2 with the R396Q mutation, and 1 with deletion of GATA2 locus), 4 of 10 patients in Kazenwadel et al (T354M, 2 distinct large deletions of GATA2 locus and a frameshift starting from L332),10 6 of 28 patients in Hahn et al (T354M or deletion of T355),7 and the case described by Bodor et al (T354M),9 suggesting that the monosomy 7q is a recurrent secondary event linked to the overt transformation. Loss of the long arm of chromosome 7 is a frequent event in MDS and AML. In addition to RUNX1 and CEBPA mutations, monosomy 7 was associated with pure familial MDS/AML in 14 pedigrees with an autosomal-dominant transmission and a young age (< 18 years).21 The different parental origins of the remaining chromosome 7 within the same family led to the hypothesis of the existence of a “mutator gene” located outside of chromosome 7.21 GATA2 might be this putative mutator gene suggested by Minelli et al.21 GATA2 mutation could act as a preleukemic event followed by an overt transformation due to the loss of genes located in the long arm of chromosome 7.
The main genes involved in congenital neutropenia were characterized during the last decade, with ELANE being involved in half of them (for review, see Donadieu et al1 ). In the present study, we identified GATA2 mutations as a cause of congenital mild neutropenia associated with a high risk of leukemic transformation. Monocytosis is frequently associated with neutropenia.1 In marked contrast, our patients with GATA2 mutations presented with monocytopenia with a median of 0.05 G/L.
Patients of 2 of the pedigrees described herein (P3 and P5) have a syndrome related to the WHIM syndrome, although the BM myelokathexis feature was missing. They display the typical WHIM-associated dysfunctions of the CXCR4 chemokine receptor pathway without mutations of CXCR4 that are frequent in WHIM syndrome.17,18,22 The monocytopenia, together with the infectious spectrum composed by pyogenic or mycobacteria infections and HPV-related warts, have some similarities to the MonoMAC syndrome. In support of this possibility, we found that both patients harbor heterozygous mutation of GATA2. Nevertheless the potential mechanisms of pathogenesis remain largely unknown. Functional studies are needed to investigate this and the potential effects of GATA2 mutations on dysfunction of the CXCR4 pathway, which account for the panleukopenia of WHIM patients.23
Identification of GATA2 mutations is critical for the management of these patients. The serious and usually sudden infections occurring in 4 patients led to their deaths. The spectrum of infections associated with GATA2-deficient neutropenic patients encompasses bacterial and fungal infection with frequent skin or mucosal involvements and a higher risk of mycobacterial and viral infections, reflecting either the profound deficit of NK cells in MonoMAC syndrome13 or the importance of the cooperation between neutrophils and NK cells.24 The present study also highlights the frequent early myeloid transformation in patients carrying a GATA2 mutation associated with a poor outcome linked to a refractory status of disease after initiation of chemotherapy. Two patients in our series (P1, patient number 6227 and P6, patient number 7015) were treated with hematopoietic growth factors without benefits (lenograstim followed by sargramostim in one patient and filgrastim in the other).
Screening of GATA2 mutations in patients with mild chronic neutropenia associated with monocytopenia may be important. We propose limiting the use of hematopoietic growth factors and implementing intensive treatment such as allograft in patients with neutropenia and GATA2 mutations as soon as an unrelated donor has been identified. Genetic counseling should also be considered for patients with GATA2 mutations because of the high clinical penetrance and poor survival rates.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients and their families for their participation in this study; the Association 111 des Arts for its support; Michel Leporrier for collection of blood samples of P1; Cyril Broccardo for careful reading of the manuscript; and Miikka Vikkula and Ha-Long Phuoc Nguyen (Human Molecular Genetics, GEHU-DDUV, Université Catholique de Louvain, Brussels, Belgium) for the genetic cosegregation analyses of the pedigree P4 (II-1, III-2, III-3).
F.B. is supported by E-rare 2011-RARE-013-01 and is a member of the Laboratory of Excellence LERMIT supported by a grant from ANR (Investissements d′Avenir). The French registry is supported by grants from Amgen SAS, Chugai SA, GIS Maladies Rares, Institut de Veille Sanitaire, Inserm, and the Center de Référence des Déficits Immunitaires Héréditaires (CEREDIH: the French National Reference Center for Primary Immune Deficiencies, www.ceredih.fr).
Authorship
Contribution: M.P., C.B.-C., S.T., B.B., A.P., P.R., C.F., E.V.D.N., H.A.P., T.L., M.O., C.R., G.P., and J.D. provided DNA samples and clinical information; S.T., N.P., O.L., H.A.P., F.B., and E.D. performed the experiments; V.M.-D.M. and J.C. reviewed the blood and BM slides; M.P., S.T., and J.D. reviewed the clinical information; M.P., C.B.-C., S.T., J.D., and E.D. designed the research and analyzed the results; M.P., S.T., J.D., and E.D. produced the tables and figures; and J.D. and E.D. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pr Eric Delabesse, Laboratoire d'Hématologie, Pavillon Lefebvre, Hôpital Purpan, Place du Dr Baylac, TSA40031, 31059 Toulouse cedex 9, France; e-mail: delabesse.e@chu-toulouse.fr.
References
Author notes
M.P., C.B.-C., S.T., J.D., and E.D. contributed equally to this work.