Key Points
Some CD34+CD38+ intermediate hematopoietic progenitor cells express HIV-1 entry receptors and are susceptible to direct infection by HIV.
Blood progenitors from HIV-exposed, humanized BLT mice show impaired hematopoietic potential and give rise to progeny that harbor provirus.
Abstract
HIV infection has been associated with defective hematopoiesis since the earliest days of the HIV/AIDS epidemic. Generation of all hematopoietic lineages suffers in the face of infection. The mechanisms by which HIV impairs normal blood cell development remain unclear, and direct infection of intermediate hematopoietic progenitors has not been established as a source of HIV-associated hematopoietic pathology. Here, we demonstrate infection of multiple subsets of highly purified intermediate hematopoietic progenitors by wild-type HIV both in vitro and in vivo. Although direct infection is clearly cytotoxic, we find that some infected progenitors can survive and harbor proviral DNA. We report intermediate hematopoietic progenitors to be a novel target of infection and their permissivity to infection increases with development. Further, the nonobese diabetic severe combined immunodeficiency common γ chain knockout-bone marrow-liver-thymus humanized mouse provides a unique model for studying the impact of HIV infection on bone marrow–based human hematopoiesis.
Introduction
HIV is the etiologic agent responsible for AIDS. Hematopoietic abnormalities are common manifestations of systemic infection,1 and multilineage hematopoiesis clearly suffers in the face of HIV infection.2 Early in the AIDS epidemic, it was realized that HIV infection manifests in defective hematopoiesis.3,4 Although erythropoiesis and megakaryopoiesis are most impaired,5,6 the development of all hematopoietic lineages is impacted by HIV infection.7,8 The degree of hematopoietic pathology correlates with the stage of disease progression,9 and end-stage disease is characterized by pancytopenia.10 The introduction of highly active antiretroviral therapy (HAART) regimens in the mid-1990s dramatically changed many aspects of living with HIV infection, including drastic improvements in hematopoiesis.11 Although HAART clearly ameliorates HIV-associated hematosuppression, blood cell development is not completely restored.12 Moreover, long-term toxicity concerns are spurring the idea of moving treatment away from drug therapy.13
The site of human hematopoiesis in adults is the bone marrow, whereas it is the liver in the fetus.14 Historically, HIV has been thought not to penetrate these compartments; however, the bone marrow microenvironment is not isolated from virus exposure. It is subject to normal circulation and is thus exposed to infected cells and free virus of infected individuals’ blood. Moreover, a number of bone marrow–resident cells are subject to infection themselves.15 Although a litany of indirect causes of HIV-associated hematosuppression has been explored,16 it is unclear whether hematopoietic progenitors can themselves become infected by HIV-117-21 and, if so, what would be the resulting impact on hematopoiesis.
Hematopoietic progenitor cells (HPCs) comprise a diverse population and include both early and intermediate progenitors. It is generally accepted that all hematopoietic progenitors express the cell surface antigen CD34. Early and intermediate populations can be distinguished by the expression of CD38, the former being negative for this antigen. Each of these subpopulations expresses diverse and distinct sets of cell surface antigens.22,23 Intermediate progenitors include the common myeloid progenitor (CMP) that can give rise to all myeloid, erythroid, and megakaryocyte lineages; the granulocyte-monocyte progenitor (GMP); and the megakaryocyte-erythroid progenitor (MEP). Infection of an intermediate progenitor would therefore have significant consequences for multiple cell types.
The susceptibility of a cell to infection by HIV is determined by the expression of surface molecules CD4 and either the chemokine receptor CXCR4 or CCR5 that bind the HIV-1 envelope and mediate entry of the virus into the cell. Early studies found low-level expression of the necessary surface proteins on early progenitors to allow viral entry but concluded that these cells are not infected at an appreciable level.24-27 Recent work confirmed low expression of some these receptors in the earliest of hematopoietic progenitors but did not address expression on intermediate progenitors.28
Several factors confound the study of HIV-1–associated hematosuppression in patient samples. Some antiretroviral medications are known to impair hematopoiesis, whereas others are thought to alleviate hematosuppression.29,30 In addition, HIV-associated opportunistic infections and bone marrow neoplasms, as well as many of the drugs used to treat them, are known to disrupt normal hematopoiesis. An animal model that bypasses these confounding factors is required.
Consequently, we sought to resolve some of these issues and assess the impact of HIV infection of intermediate HPCs using a series of in vitro and in vivo studies in humanized mice. In the present study, we demonstrate significant pathology results from direct infection of hematopoietic progenitors by HIV in vitro. Further, we show that purified subpopulations of intermediate hematopoietic progenitors are increasingly susceptible to infection by HIV in vitro. We found a consistent phenotype in humanized nonobese diabetic (NOD) severe combined immunodeficiency (SCID) common γ chain knockout (NSG) mice that have been implanted with human fetal bone marrow-liver-thymus tissue (BLT), which have been exposed to HIV. In the absence of drug treatment, bone marrow–derived HPCs are susceptible to infection, which results in hematosuppression. We go on to establish that these HPCs can give rise to progeny that harbor proviral DNA.
Methods
Cells and cell sorting
Human HPCs were obtained from either cord blood or fetal liver obtained with institutional review board approval. CD34+ HPCs were isolated from the buffy coat after Ficoll-Hypaque gradient centrifugation by magnetic associated cell sorting (MACS) using CD34+ direct magnetic beads (Miltenyi Biotec, Auburn, CA) and MACS LS or MS separation columns (Miltenyi Biotec) or fluorescent-activated cell sorting (FACS) where indicated. To increase purity, each sample was passed over 2 columns when MACS was used. Depletion of CD3-, CD14-, and CD16-expressing cells was performed using purified murine antibodies conjugated to goat anti-mouse magnetic beads (Miltenyi Biotec) and then each sample was passed over 2 LS columns (Miltenyi Biotec) on magnets. The flow through was enriched for CD34+ cells using direct CD34 magnetic beads (Miltenyi Biotec); each sample was then passed over 2 LS columns. Released cells were stained with anti-CD38 phycoerythrin (PE), anti-CD45RA fluorescein isothiocyanate, and anti-CD110 allophycocyanin (APC) antibodies for FACS performed at the Broad Stem Cell Institute Flow Cytometry Core. The CMP was defined as CD34+ CD38+ CD45RA− CD110−, the GMP as CD34+ CD38+ CD45RA+ CD110−, and the MEP as CD34+ CD38+CD110+ (Table 1).
Cells were maintained in StemSpan serum-free medium (Stem Cell Technologies, Vancouver, BC, Canada) supplemented with stem cell factor (Invitrogen, Grand Island, NY) (100 ng/mL), thrombopoietin (Invitrogen) (100 ng/mL), FMS-like tyrosine kinase 3 ligand (Invitrogen) (100 ng/mL), and penicillin/streptomycin in a 5% CO2 37°C incubator. For the mouse experiment, marrow was flushed from murine femur, passed through a 100-μm mesh filter, and washed twice with phosphate-buffered saline (PBS). Human CD34+ cells were then isolated by the method described previously.
Vector and wild-type virus growth and infections
The NL4HSA-eGFP vector was prepared as previously described.31 Briefly, the nucleotide sequence encoding the murine heat-stable antigen (HSA) was inserted into the vpr sequence and the nucleotide sequence encoding the enhanced green fluorescent protein (eGFP) was inserted into the viral envelope sequence. To create infectious virion, the vector was complemented with the envelope of the vesicular stomatitis virus type-G (VSV-G) and the ∆8.2 packaging construct by liposome-mediated (Lipofectamine 2000; Invitrogen) transfection of 293FT human embryonic kidney cells.
Wild-type viruses were created similarly from proviral plasmids p89.6, pYJRCSF, and pNL4-3. Supernatants were harvested 48 hours later, passed through a 45-μm vacuum filter, and stored at −80°C until use. Infections of primary cells were performed on a rocking platform for 2 hours in a 37°C 5% CO2 incubator. Cells were then pelleted and washed twice with PBS and cultured as described above for 48 hours, when they were harvested for further analysis.
Flow cytometry and antibodies
The following were used: anti-CD4 ECD (clone GK1.5; Beckman Coulter, Brea, CA), anti-CCR5 PerCP-Cy5.5 (clone 3A9; BD Pharmingen, San Jose, CA), anti-CXCR4 PE-Cy7 (clone 12G5; BD Pharmingen), anti-CD34 APC (clone 8G12; Beckman Coulter), anti-CD38 PE (clone LS198-4-3; Beckman Coulter), anti-CD45RA fluorescein isothiocyanate (clone HI100; BD Pharmingen), anti-CD123 e450 (clone 6H6; eBiosciences, San Diego, CA), anti-CD110 APC (clone 167639; R&D Systems), anti-CD34 PE (clone Immu133; Beckman Coulter), pure anti-CD3 (clone HIT3a; BD Pharmingen), pure anti-CD14 (clone M5E2; BD Pharmingen), and pure anti-CD16 (clone 3G8; BD Pharmingen). Stained cells were analyzed on a Becton Dickson Fortessa at the Broad Stem Cell Research Center Flow Cytometry Core Laboratory at the University of California Los Angeles. All flow cytometry data were analyzed with FlowJo software.
Colony-forming assays
Cells were seeded in complete methylcellulose (Methocult H04034; Stem Cell Technologies) containing azidothymidine (AZT) 10 μM and Indinavir 100 nM at a concentration of 1000 cells/mL and plated in 35-mm grid plates, 1 mL/plate, in triplicate per mouse. When examining megakaryopoiesis, cells were plated at a density of 5000 cells/mL in semisolid collagen-based medium (Megacult-C; Stem Cell Technologies). Colonies were counted 2 weeks later in a blinded fashion using a Nikon Eclipse TE300 microscope at a magnification of ×400. Discrete, individual colonies were picked with a with a pipette tip, washed 3 times in 500 μL of sterile PBS, and stored at −80°C until DNA extraction.
Analysis of viral DNA
Detection of viral DNA was accomplished through the use of quantitative real-time polymerase chain reaction (qRT-PCR) as previously described.32 DNA was isolated from cells by standard phenol-chloroform extraction followed by ethanol precipitation. Primers BGF10 and BGR1 as well as probe BGX1 were used to amplify cellular β-globin, which we used as an internal standard. Primers SR1 and 661 along with probe ZXF were used to amplify the Gag/LTR junction, which indicates full-length viral DNA. Known quantities of standards were run in parallel, creating a standard curve for both HIV and β-globin, and sample DNA was quantified by extrapolation from the standard curve. Samples were amplified using an IQCycler (Bio-Rad, Hercules, CA) system and analysis was done with iQ5 software (Bio-Rad). Analysis for integration site was carried out as previously described.33 Briefly, genomic DNA was fragmented and integrated viral DNA annealed to the biotinylated primer BNL9005F for enrichment mediated by streptavidin-coated magnetic beads. Isolated DNA was amplified by a 2-step, nested PCR protocol. The first step used primers NL495F and NL9560R. The second nested PCR step amplified DNA with primers NL510F and NL94533R. Products resulting from the second step were cloned and sequenced.
Mice
All work involving mice was carried out under the approval of the University of California Los Angeles Animal Research Committee. NSG-BLT humanized mice were constructed as previously described.34 Briefly, after sublethal irradiation, NSG mice were implanted with pieces of human fetal liver and thymus beneath the kidney capsule and injected with allogeneic CD34+ cells to seed the bone marrow. Four weeks later, mice were analyzed for engraftment of human cells and immune reconstitution by flow cytometric analysis of peripheral blood. Cells were stained with anti-CD45, anti-CD3, anti-CD4, and anti-CD8 antibodies. Once engraftment of human cells was confirmed, 150 ng p24 HIV was delivered by retro-orbital injection.
Statistics
Data were analyzed with paired or unpaired Student t test as indicated. All statistical analyses were performed with GraphPad Prism v6.0 (GraphPad Software). Values with P < .05 were considered significant.
Results
Direct infection of CD34+ HPCs impacts multiple hematopoietic lineages
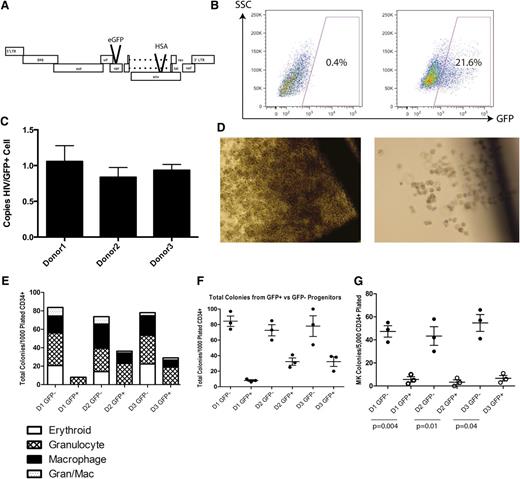
We first investigated whether direct infection in vitro of a mixed population of CD34+ HPCs derived from fetal liver would diminish their hematopoietic potential. A VSV-G pseudotyped, replication-deficient vector based on the NL4-3 strain of HIV was used to infect fetal liver–derived CD34+ HPC from 3 donors. This NL4HSA-eGFP vector is deficient in the viral genes vpr and env because of insertions of the coding sequences for the murine HSA and eGFP, respectively (Figure 1A). Forty-eight hours after infection, cells were harvested and sorted by FACS to high purity for the presence and absence of GFP expression before being plated in complete methylcellulose to allow differentiation (Figure 1B). A fraction of these cells was analyzed by quantitative PCR to determine the number of integrants in each GFP-positive cell. On average, each cell contained approximately 1 integrant (Figure 1C).
VSV-G pseudotyped infection of CD34+ HPCs results in developmental pathology in multiple hematopoietic lineages. (A) The NL4HSA-eGFP vector is based on the NL4-3 strain of HIV-1. The vector is deficient in env and vpr because of the insertion of marker genes and was pseudotyped with VSV-G to produce viral vector particles. (B) FACS strategy for differentiating GFP-positive and GFP-negative CD34+ hematopoietic progenitor cells 48 hours after transduction. (C) Quantitative PCR analysis for copies of HIV per GFP+ cell indicates a single integrant on average for each donor. (D) Colonies grown in methylcellulose derived from NL4HSA-eGFP–transduced cells (right) are in general much smaller than those derived from nontransduced cells (left). (E) CD34+ hematopoietic progenitors transduced with NL4-HSA-eGFP generate far fewer colonies than do nontransduced cells. Each mark represents an individual methylcellulose plate and the data represent the mean ± the standard error of the mean (SEM). (F) There is a skewing of lineages in NL4-HSA-eGFP–transduced cultures compared with nontransduced. Erythroid colonies are particularly impacted. (G) Megakaryocyte colonies are likewise diminished from transduced progenitors. Each mark represents an individual plate and the data represent the mean ± SEM. LTR, long terminal repeat; SSC, side scatter.
VSV-G pseudotyped infection of CD34+ HPCs results in developmental pathology in multiple hematopoietic lineages. (A) The NL4HSA-eGFP vector is based on the NL4-3 strain of HIV-1. The vector is deficient in env and vpr because of the insertion of marker genes and was pseudotyped with VSV-G to produce viral vector particles. (B) FACS strategy for differentiating GFP-positive and GFP-negative CD34+ hematopoietic progenitor cells 48 hours after transduction. (C) Quantitative PCR analysis for copies of HIV per GFP+ cell indicates a single integrant on average for each donor. (D) Colonies grown in methylcellulose derived from NL4HSA-eGFP–transduced cells (right) are in general much smaller than those derived from nontransduced cells (left). (E) CD34+ hematopoietic progenitors transduced with NL4-HSA-eGFP generate far fewer colonies than do nontransduced cells. Each mark represents an individual methylcellulose plate and the data represent the mean ± the standard error of the mean (SEM). (F) There is a skewing of lineages in NL4-HSA-eGFP–transduced cultures compared with nontransduced. Erythroid colonies are particularly impacted. (G) Megakaryocyte colonies are likewise diminished from transduced progenitors. Each mark represents an individual plate and the data represent the mean ± SEM. LTR, long terminal repeat; SSC, side scatter.
After 14 days, the size of colonies in the cultures derived from GFP+ progenitors was significantly smaller than those derived from GFP− progenitors (Figure 1D). Further, the total colony number was much lower in GFP+ derived cultures. Although the average total number of colonies per plate from uninfected cells from 3 separate donors was 78 ± 6, it is 24 ± 14 for infected cells, a statistically significant difference in a paired Student t test (P = .04) (Figure 1E).
Notably, erythroid and megakaryocyte development are particularly impacted. No erythroid colonies developed from infected cells, whereas the mean number from uninfected progenitors was 19 ± 3 (P = .02 paired t test). The numbers of granulocyte colonies and mixed granulocyte/macrophage colonies were not statistically different between the 2 cell populations. Interestingly, macrophage colonies also showed significant differences between conditions: the mean number of colonies derived from GFP+ cells was 6 ± 3 compared with 71 ± 3 (P = .008) in colonies derived from GFP− progenitors (Figure 1F). Megakaryocytes require a different, collagen-based medium for growth and so are not seen in the same cultures with myeloid or erythroid cells. When examined for megakaryopoietic potential, that of the GFP+ cells was severely impaired when compared with GFP− cells: the mean number of colonies derived from GFP+ cells was 5 ± 1 compared with 48 ± 3 from GFP− progenitors (P = .003) (Figure 1G). These results indicate that direct infection by HIV is toxic to multilineage hematopoiesis.
Intermediate HPCs express cell surface receptors necessary for HIV-1 entry
The CD34+ HPC population is diverse and includes the early progenitors long-term repopulating hematopoietic stem cell (HSC), short-term repopulating HSC, and multipotent progenitor in addition to the intermediate progenitors CMP, GMP, and MEP (Figure 2A). We next analyzed the intermediate HPCs to determine expression of proteins required for HIV entry. Both cord blood and fetal liver–derived CD34+ HPCs from 3 donors each were analyzed by flow cytometry for the presence of CD4, CXCR4, and CCR5 in conjunction with markers that define each different progenitor population. Within the CD34+38+ population, CD45RA and the α chain of the interleukin-3 receptor (CD123) were used to distinguish different intermediate progenitors (Figure 2B). The MEP is negative for both, the CMP is positive for CD123, and the GMP is positive for CD45RA (Table 1).22
CD34+CD38+ intermediate hematopoietic progenitors express cell surface receptors necessary for HIV-1 entry. (A) Within the CD34+ HPC population, CD38 coexpression indicates a more mature phenotype and includes the CMP, MEP, and GMP. (B) CD34+ cells were enriched from fetal liver and assayed by flow cytometry for the presence of HIV entry receptors in conjunction with definitive surface markers of intermediate hematopoietic progenitors. CD34+CD38+ double-positive cells were gated (left) to select the CMP, GMP, and MEP based on CD123 and CD45RA expression (right). (C) Each population of intermediate hematopoietic progenitor was examined for CD4 and CCR5 expression by flow cytometry. (D) These same populations were also examined for CD4 and CXCR4 expression by flow cytometry. LT-HSC, long-term hematopoietic stem cell; MPP, multipotent progenitor; ST-HSC, short-term hematopoietic stem cell.
CD34+CD38+ intermediate hematopoietic progenitors express cell surface receptors necessary for HIV-1 entry. (A) Within the CD34+ HPC population, CD38 coexpression indicates a more mature phenotype and includes the CMP, MEP, and GMP. (B) CD34+ cells were enriched from fetal liver and assayed by flow cytometry for the presence of HIV entry receptors in conjunction with definitive surface markers of intermediate hematopoietic progenitors. CD34+CD38+ double-positive cells were gated (left) to select the CMP, GMP, and MEP based on CD123 and CD45RA expression (right). (C) Each population of intermediate hematopoietic progenitor was examined for CD4 and CCR5 expression by flow cytometry. (D) These same populations were also examined for CD4 and CXCR4 expression by flow cytometry. LT-HSC, long-term hematopoietic stem cell; MPP, multipotent progenitor; ST-HSC, short-term hematopoietic stem cell.
In a representative flow cytometry assay, some populations of multipotent progenitors were detected that coexpress CD4 and CXCR4 or CCR5. In cord blood–derived cells, 10% of GMPs coexpress CD4 and CCR5 (Figure 2C), whereas 14% coexpress CD4 and CXCR4 (Figure 2D). In the CMP population, approximately 3% coexpress CD4 and CCR5 and almost 7% coexpress CD4 and CXCR4. MEPs appear to express viral coreceptors but they are not coexpressed with CD4. These data are consistent in both cord blood and fetal liver–derived HPCs and there are no significant differences in the expression of HIV entry receptors when comparing the HPC populations between both tissue types (Table 2). Therefore, some intermediate hematopoietic progenitors express detectable levels of the necessary cell surface receptors to allow viral entry.
Susceptibility of intermediate hematopoietic progenitors to HIV-1 infection in vitro
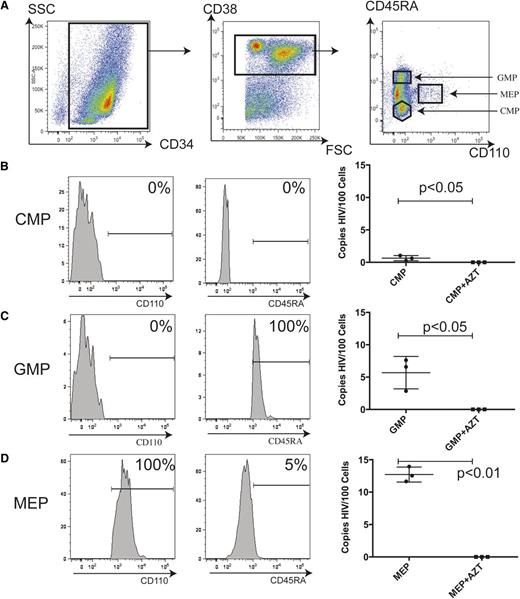
To determine if these patterns and levels of receptor expression are sufficient to render intermediate hematopoietic progenitors susceptible to infection by HIV-1, fetal liver–derived CD34+ HPC from 3 donors were highly purified by FACS (Figure 3A). The CMP, GMP, and MEP populations were purified based on CD38, CD45RA, and CD110 (the thrombopoietin receptor) expression (Figure 3A-D, left and center). Here, we chose to use CD110 instead of CD123 to promote higher purity based on previously published results.35 To prevent contamination with differentiated cells, samples were first depleted of CD3, CD14, and CD16, thereby eliminating T-cell and macrophage populations (supplemental Figure 1). Sorted cells were infected with HIV89.6, a viral strain capable of using either CXCR4 or CCR5 coreceptor. qRT-PCR analysis detected infection in the CMP population at a rate of 1 in 100 cells (P = .04) (Figure 3B, right). GMPs were infected at an average rate of 6 ± 1% (P = .02) (Figure 3C, right). Strikingly, the MEP shows a 13 ± 1% (P < .0001) infection rate (Figure 3D, right). Cells cultured in the presence of the reverse transcriptase inhibitor AZT were negative for viral DNA. These data indicate that different populations of HPCs show increasing susceptibility to infection with progressive hematopoietic development.
Intermediate hematopoietic progenitors are increasingly permissive to HIV-1 infection in vitro with progressive development. (A) CD34+ cells were sorted to high purity by MACS from mononuclear cells purified from human fetal liver. Flow cytometry shows that of the live sorted cells, 98% express CD34 (left). From these, CD38+ cells were sorted to isolate each subpopulation of intermediate hematopoietic progenitors (center). The CMP, MEP, and GMP are all CD34+CD38+; the CMP expresses neither CD45RA nor CD110. Only the MEP expresses CD110; the GMP is CD45RA+CD110− (left). (B) Purity of the sorted CMP is indicated by CD110 expression (left) and CD45RA expression (center). Cells were infected with HIV89.6 and assayed by qRT-PCR 48 hours later for full-length viral DNA (right) in the presence or absence of AZT. Each mark indicates a single donor; the data represent the mean ± SEM. (C) Purity of sorted GMP indicated by CD110 expression (left) and CD45RA expression (center). (D) Purity of the sorted MEP indicated by CD110 expression (left) and CD45RA expression (center). FSC, forward scatter; SSC, side scatter.
Intermediate hematopoietic progenitors are increasingly permissive to HIV-1 infection in vitro with progressive development. (A) CD34+ cells were sorted to high purity by MACS from mononuclear cells purified from human fetal liver. Flow cytometry shows that of the live sorted cells, 98% express CD34 (left). From these, CD38+ cells were sorted to isolate each subpopulation of intermediate hematopoietic progenitors (center). The CMP, MEP, and GMP are all CD34+CD38+; the CMP expresses neither CD45RA nor CD110. Only the MEP expresses CD110; the GMP is CD45RA+CD110− (left). (B) Purity of the sorted CMP is indicated by CD110 expression (left) and CD45RA expression (center). Cells were infected with HIV89.6 and assayed by qRT-PCR 48 hours later for full-length viral DNA (right) in the presence or absence of AZT. Each mark indicates a single donor; the data represent the mean ± SEM. (C) Purity of sorted GMP indicated by CD110 expression (left) and CD45RA expression (center). (D) Purity of the sorted MEP indicated by CD110 expression (left) and CD45RA expression (center). FSC, forward scatter; SSC, side scatter.
HPCs from HIV-infected humanized mice are impaired in hematopoietic potential
To model the impairment of hematopoiesis seen in HIV-1–infected patients, we chose to employ the NSG-BLT humanized mouse. Mice were infected with 1 of 3 strains of HIV that are distinguished by their cellular coreceptor use: HIVNL4-3, HIV89.6, and HIVJRCSF. HIVNL4-3 makes use of CXCR4, whereas HIVJRCSF requires CCR5 for entry (Figure 4A). Two months after infection, infection was confirmed by the presence of proviral DNA in the peripheral blood mononuclear cells of infected mice (Figure 4B). Cells isolated from the bone marrow of 3 mice exposed to HIVNL4-3 were analyzed by flow cytometry and compared with unexposed mice for the presence of viral protein expression in intermediate HPC populations. Representative flow plots indicate that HIV GAG protein was clearly detected in the MEP (Figure 4C-D). Although there were no clear indications of protein expression in the CMP or GMP, on average 3.65% ± 0.63% of the MEP cells were positive for GAG expression. Human CD34+ cells were then isolated from the bone marrow and enriched by MACS to high purity (Figure 4E). When analyzed by qRT-PCR, infection could be detected in the CD34+ cells from each of the mice exposed to virus. Cells isolated from the bone marrow of mice infected with any of the 3 strains all showed between 6 and 9 copies of viral DNA per 1000 cells (Figure 4F). Therefore, a significant fraction of human CD34+ cells can be infected with various strains of HIV in vivo, regardless of viral tropism.
Intermediate HPCs derived from the bone marrow of HIV-infected NSG-BLT mice are productively infected as shown by HIV-GAG expression. (A) Schematic of experimental design for infecting humanized NSG-BLT mice and isolating CD34+ HPCs from their bone marrow. (B) Peripheral blood mononuclear cells from infected mice were analyzed by PCR for the presence of full-length viral DNA relative to the number of copies of human β-globin to determine the percent of infected cells. Each mark represents the average of 3 reactions for each mouse; the total data represent the pooled mean ± SEM of all reactions from all mice. Cells isolated from the murine bone marrow were analyzed for the presence of human intermediate HPC markers in conjunction with HIV-GAG expression. (C) Cells derived from uninfected mice. (D) Cells derived from mice exposed to HIVNL4-3. (E) Representative flow cytometric analysis of CD34+ purity after sorting from BLT bone marrow. (Left) Isotype control; (right) CD34 staining at 98.5% purity. (F) Human CD34+ HSC derived from BLT bone marrow harbor full-length HIV DNA when analyzed by quantitative PCR. Each mark indicates the average of triplicate assays in a single mouse; the total data represent the pooled mean ± SEM of all reactions from all mice. The dashed line indicates the limit of detection of the assay. JRCSF, the patient code used to define the particular strain of HIV.
Intermediate HPCs derived from the bone marrow of HIV-infected NSG-BLT mice are productively infected as shown by HIV-GAG expression. (A) Schematic of experimental design for infecting humanized NSG-BLT mice and isolating CD34+ HPCs from their bone marrow. (B) Peripheral blood mononuclear cells from infected mice were analyzed by PCR for the presence of full-length viral DNA relative to the number of copies of human β-globin to determine the percent of infected cells. Each mark represents the average of 3 reactions for each mouse; the total data represent the pooled mean ± SEM of all reactions from all mice. Cells isolated from the murine bone marrow were analyzed for the presence of human intermediate HPC markers in conjunction with HIV-GAG expression. (C) Cells derived from uninfected mice. (D) Cells derived from mice exposed to HIVNL4-3. (E) Representative flow cytometric analysis of CD34+ purity after sorting from BLT bone marrow. (Left) Isotype control; (right) CD34 staining at 98.5% purity. (F) Human CD34+ HSC derived from BLT bone marrow harbor full-length HIV DNA when analyzed by quantitative PCR. Each mark indicates the average of triplicate assays in a single mouse; the total data represent the pooled mean ± SEM of all reactions from all mice. The dashed line indicates the limit of detection of the assay. JRCSF, the patient code used to define the particular strain of HIV.
To determine if human hematopoietic progenitors derived from infected mice show impaired hematopoiesis, human CD34+ cells were isolated from bone marrow and plated in complete methylcellulose supplemented with the reverse transcriptase inhibitor AZT and the protease inhibitor indinavir to prevent viral spread within the culture. Two weeks later, plates from mice infected with HIV showed drastically fewer colonies than those from uninfected mice. HPCs isolated from uninfected mice consistently produce, on average, more than 60 colonies per 1000 CD34+ cells plated, whereas CD34+ HPCs derived from infected mice produce, on average, less than 20 colonies per 1000 CD34+ HPCs (Figure 5A). Generation of all lineages except granulocytes was depressed in cultures from infected mice (Figure 5B). Notably, the percentage of erythroid colonies was strikingly lower in cultures derived from infected mice. HPCs derived from uninfected mice produced, on average, almost 30% erythroid colonies, whereas HPCs from infected mice yielded fewer than 15% erythroid colonies (Figure 5C). Thus, normal hematopoiesis suffers considerably in the presence of HIV-1 infection.
HPCs from HIV-infected NSG-BLT mice generate colonies that harbor proviral DNA. (A) The total number of colonies that develops from each infected mouse is significantly less than those from uninfected mice. Each mark represents the average of triplicate assays in a single mouse; the total data represent the pooled mean ± SEM from all mice. (B) Phenotypic analysis of colonies indicates significant differences in lineage commitment from HSC derived from infected mice compared with uninfected mice. (C) Erythroid lineage development is particularly impaired in the HSC derived from HIV-1–infected mice. Each mark represents the average of triplicate assays in a single mouse; the total data represent the pooled mean ± SEM from all mice. (D) Colonies derived from human CD34+ cells isolated from the bone marrow of infected and uninfected mice were assayed by qRT-PCR for the presence of full-length viral DNA. Colonies were first phenotyped visually by light microscopy. E, erythroid; G, granulocyte; GM, granulocyte-macrophage mixed; M, macrophage. Percentages are of that particular colony type in which HIV was detected compared with the total number of that colony type assayed. (E) HIV+ colonies were assayed by alu-Gag PCR to determine integration site. Following successful amplification, amplicons were sequenced to determine the gene into which the virus integrated. Four different mice were assayed, representing infection by each of the 3 viral strains.
HPCs from HIV-infected NSG-BLT mice generate colonies that harbor proviral DNA. (A) The total number of colonies that develops from each infected mouse is significantly less than those from uninfected mice. Each mark represents the average of triplicate assays in a single mouse; the total data represent the pooled mean ± SEM from all mice. (B) Phenotypic analysis of colonies indicates significant differences in lineage commitment from HSC derived from infected mice compared with uninfected mice. (C) Erythroid lineage development is particularly impaired in the HSC derived from HIV-1–infected mice. Each mark represents the average of triplicate assays in a single mouse; the total data represent the pooled mean ± SEM from all mice. (D) Colonies derived from human CD34+ cells isolated from the bone marrow of infected and uninfected mice were assayed by qRT-PCR for the presence of full-length viral DNA. Colonies were first phenotyped visually by light microscopy. E, erythroid; G, granulocyte; GM, granulocyte-macrophage mixed; M, macrophage. Percentages are of that particular colony type in which HIV was detected compared with the total number of that colony type assayed. (E) HIV+ colonies were assayed by alu-Gag PCR to determine integration site. Following successful amplification, amplicons were sequenced to determine the gene into which the virus integrated. Four different mice were assayed, representing infection by each of the 3 viral strains.
Interestingly, colonies derived from mice infected by each of the 3 strains showed presence of full-length viral DNA by qRT-PCR. Approximately half of the colonies assayed from HIVNL4-3- or HIV89.6-infected mice were positive for viral DNA. Cultures derived from HIVJRCSF-infected mice were 14% positive for viral DNA. All 3 viruses infected precursors of the erythroid lineage at a high rate (Figure 5D). We went on to analyze the integration of the proviral DNA found in these colonies. Of the 4 infected colonies tested, each showed only 1 or 2 integration sites (Figure 5E), thus eliminating the possibility of contamination by exogenous genomic DNA. Because each colony likely arises from a single cell and viral spread was blocked by antiretroviral drugs, coupled with the presence of only limited integration sites, our data demonstrate that intermediate hematopoietic progenitors can become infected and give rise to stably infected progeny.
Discussion
As the HIV-1 positive population lives longer with infection, this aging demographic is a growing area of interest. Anemia and thrombocytopenia have the potential to present significant mortality and morbidity in this group as they face increasing susceptibility to age-related illnesses. HIV-1 infection of stem and progenitor cells has been proposed since the earliest days of research on the virus, but the significance remains under debate.
We have shown, using a pseudotyped reporter virus, that when a CD34+ HPC becomes infected, severe developmental pathology results in all hematopoietic lineages, independent of entry or binding of viral envelope to the cell surface. It is interesting that some lineages are more impaired than others in this assay. We speculate that development to specific lineages may impart increased susceptibility to infection-associated cytopathology, as postulated by Carter et al17 ; however, this requires further study to determine the underlying mechanism. We go on to demonstrate that some subsets of intermediate HPCs do indeed express HIV-1 entry receptors. We show here for the first time that both the GMPs and the MEPs are targets of direct infection by wild-type HIV-1 in vitro in highly purified populations of intermediate hematopoietic progenitors. Our finding that the MEP population shows relatively high susceptibility to infection despite the lack of detectable viral entry receptor coexpression is interesting. Although there are reports of CD4-independent entry by HIV-1,36 the mechanism of viral entry into each HPC progenitor population demands further study.
We then show that intermediate hematopoietic progenitors are susceptible to direct infection in an in vivo system. In the context of unmitigated infection in the humanized NSG-BLT mouse, bone marrow–derived human CD34+ HPCs were defective in their hematopoietic potential. Further, quantitative PCR analysis showed that a significant number of the colonies derived from these HPCs harbor proviral DNA. Less than 0.1% of CD34+ cells derived from uninfected mice give rise to colonies in methylcellulose, thus there is a high degree of selection for the ability to form colonies ex vivo. The relatively high rate of infection demonstrated in the resulting colonies may be due to an increased susceptibility of active intermediate progenitors to viral infection relative to other CD34+ cells. Even though our laboratory previously demonstrated defective hematopoiesis using progenitor cells isolated from the combined thymus/liver (Thy/Liv) implants in a SCID-human mouse model, we were unable to document direct infection of progenitors in the Thy/Liv implant.37 Our current in vivo model is more physiologically relevant because the infection is systemic and the human CD34+ cells are isolated from the bone marrow, as opposed to the Thy/Liv implant in the SCID-human mouse. Together, these results underscore the utility of the NSG-BLT model to study the impact of HIV infection on human hematopoiesis.
Although it has been shown that some populations of progenitors can become infected with reporter viruses in vitro,38 this has not been demonstrated with wild-type HIV nor in an animal model system. Recent work by others shows that HIV-1 DNA could not be detected in highly purified bone marrow CD34+ cells derived from patients.19,20 In these studies, all patients were on long-term, suppressive pharmacotherapy. Because of their relatively short lifespan,39 we do not believe intermediate progenitors represent a stable viral reservoir. However, intermediate progenitors could potentially be a dynamic reservoir maintained by low-level viremia in the absence of HAART and as such may warrant further study as a novel target for antiviral therapy.
This is the first demonstration of significant infection of hematopoietic progenitors by CCR5 tropic virus, albeit much less than that of CXCR4 or dual tropic virus. Likewise, this is the first demonstration of in vivo infection by wild-type HIV of bone marrow–derived human CD34+ in a nonprimate animal model. We also show that these progenitors give rise to progeny that survive infection and harbor proviral DNA. It remains to be determined whether the infection we observe in cells derived from infected HPC is productive. Nonetheless, our studies conclusively establish the potential for infection of intermediate HPCs by HIV-1 in vivo.
These results have important implications for potential therapeutics aimed at maintaining and improving the health of HIV-infected individuals. They indicate that specific protection of the MEP from infection is necessary in order to protect megakaryo-erythropoiesis and restore normal hematopoiesis in HIV-1–positive individuals. Blocking or reversing the process of HIV-associated bone marrow cytopenia would improve the health of those living with infection and could potentially ameliorate some of the challenges associated with treating HIV-associated lymphomas and end-stage AIDS hematopoietic crisis.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank H. J. Brown for her critical assessment of the manuscript. The following reagents were obtained from the National Institutes of Health (NIH) AIDS Reagent program: pYJRCSF, pNL4-3, p89.6, azidothymidine, and Indinavir. Virological measures (p24), cord blood, and peripheral blood mononuclear cells derived from anonymous donors were obtained from the University of California Los Angeles (UCLA)/Center for AIDS Research (CFAR) Virology Core Laboratory; mouse work was performed with the assistance of the UCLA/CFAR Humanized Mouse Core Laboratory.
This work was funded in part by grants from NIH/Virology and Gene Therapy Training (T32AI060567) (C.C.N.); California Institute for Regenerative Medicine Bridges to Stem Cell Research (TB1-01183) (S.N.R.); NIH/National Institute on Drug Abuse (R21DA031036-01A1) (D.N.V.), R01AI103385, and R01AI070010 (J.A.Z.); UCLA CFAR (AI028697); and the Collaboratory of AIDS Researchers for Eradication (U19 AI096113).
Authorship
Contribution: C.C.N., C.H.U., and J.A.Z. wrote the text; C.C.N., D.N.V., and J.A.Z. designed the experiments; and C.C.N., D.N.V., S.N.R., D.D., and S.G.K. performed the experiments.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jerome A. Zack, University of California Los Angeles, Department of Medicine, Division of Hematology/Oncology/Microbiology, Immunology, and Molecular Genetics, Box 957363, 190A Biomedical Science Research Building, Los Angeles, CA 90095; e-mail: jzack@ucla.edu.