Key Points
Multiple enhancers identified at the Ikzf1 locus with shared and distinct epigenetic and transcriptional properties.
Transcription factor networks that distinguish between LMPP-specific and T cell–specific Ikzf1 enhancers.
Abstract
Ikaros is a critical regulator of lymphocyte development and homeostasis; thus, understanding its transcriptional regulation is important from both developmental and clinical perspectives. Using a mouse transgenic reporter approach, we functionally characterized a network of highly conserved cis-acting elements at the Ikzf1 locus. We attribute B-cell and myeloid but not T-cell specificity to the main Ikzf1 promoter. Although this promoter was unable to counter local chromatin silencing effects, each of the 6 highly conserved Ikzf1 intronic enhancers alleviated silencing. Working together, the Ikzf1 enhancers provided locus control region activity, allowing reporter expression in a position and copy-independent manner. Only 1 of the Ikzf1 enhancers was responsible for the progressive upregulation of Ikaros expression from hematopoietic stem cells to lymphoid-primed multipotent progenitors to T-cell precursors, which are stages of differentiation dependent on Ikaros for normal outcome. Thus, Ikzf1 is regulated by both epigenetic and transcriptional factors that target its enhancers in both redundant and specific fashions to provide an expression profile supportive of normal lymphoid lineage progression and homeostasis. Mutations in the Ikzf1 regulatory elements and their interacting factors are likely to have adverse effects on lymphopoiesis and contribute to leukemogenesis.
Introduction
Lymphocyte differentiation is dependent on nuclear factors acting as key regulatory nodes that control gene expression in a cell type– and stage-specific manner. A critical node in the lymphoid lineage regulatory circuit is the Ikaros family of zinc-finger DNA binding proteins, inactivation of which causes lymphocyte disorders and lymphoid malignancies.1,2
The first major role played by Ikaros proteins is manifested in the lymphoid-primed multipotent progenitor (LMPP).3-5 Ikaros’ affiliation with a higher-order epigenetic complex that contains disparate chromatin remodeling activities on one hand primes an early lymphoid lineage transcriptional signature, whereas on the other, it represses a stem cell–specific transcriptional signature.6 Ikaros-deficient LMPPs are unable to undergo lymphoid differentiation. Instead they differentiate into the myeloid lineage while retaining significant stem cell–specific gene expression.4,6 Importantly, restriction of a hematopoietic stem cell (HSC) into an LMPP is underscored by an increase in Ikaros expression.
The second major role played by Ikaros is at stages of T- and B-cell differentiation that express high levels of Ikaros mRNA and protein.1,7 Reduction in Ikaros levels in double positive thymocytes and in pre-B cells is associated with aberrant differentiation and leukemic transformation in both mice and humans.8-15
Here, we evaluate the transcriptional mechanisms that control Ikzf1 expression in the hematopoietic system. Using a mouse transgenic reporter approach, we establish the activities of 10 lymphoid-specific clusters of DNase I hypersensitivity sites (DHSs) previously mapped at the Ikzf1 locus.16 Our studies reveal that transcription of the Ikzf1 locus is regulated by a network of epigenetic and transcription factors working together in a unique and redundant fashion to provide the appropriate levels of Ikaros expression needed for lymphocyte development.
Material and methods
Mice
Ikaros-green fluorescent protein (GFP) reporter lines (C57BL/6 × C3H) and Ikzf1 bacterial artificial chromosome (Ik-BAC) transgenic lines (C57BL/6 × C3H) were generated in the Massachusetts General Hospital transgenic core facility. Mice were bred and maintained under specific pathogen-free conditions in the animal facility in Massachusetts General Hospital, Building 149. The mice were 4 to 12 weeks old at the time of analyses.
Generation of Ikaros-GFP reporter transgenic mice
All GFP transgenic reporters used in this study are based on the B-p-GFP.16 Genomic regions J, D, E, F, G, H, and I (described in Table 1) were PCR amplified from an Ikzf1 locus containing BAC vector (Ik-BAC-3; Genome Systems, Inc., St. Louis, MO) using specific primers described in supplemental Table 5 on the Blood Web site. More detailed strategies for vector constructions and generation of mice are described in supplemental Methods.
Generation of BAC transgenic mice
Detailed strategies for construction of Ik-BAC-hCD2, Ik-BAC-lDl, and Ik-BAC-ΔD vectors, generation of mice, screening for transgene positive founders, and copy number determination are described in supplemental Methods (see also “Constructs for recombination templates for BAC engineering”).
Flow cytometric analysis
Antibodies, their specific clones, and flow cytometers (BD) used are described in supplemental Methods. Cell sorting was performed on a Moflo (Dako Cytomation). Data analysis was performed using FlowJo software (Tree Star, Inc.).
GFP reporter expression analysis in the brain
Brains were isolated from P0 embryos and photographed in whole mount in the green fluorescence channel. The samples were cleaved at the posterior edge of the basal ganglia and fixed overnight in 4% paraformaldehyde in PBS at 4°C and then dehydrated through graded sucrose, embedded in OCT, and cryosectioned.
Chromatin immunoprecipitation sequencing analysis
Chromatin immunoprecipitation sequencing (ChIP-seq) for RNA polymerase II (RNApII) CTD phospho Ser5, HEB, H3K4me1, me2, me3, me36, and H3K9Ac in wild-type thymocytes was previously described.10,17 Briefly, 107 to 108 thymocytes were formaldehyde-fixed and sonicated to an average size of 300 bp. ChIP-seq for T cell factor 1 (TCF-1) (a kind gift of Dr Kawamoto), RNApII CTD phospho Ser2 (ab5095), and the unphosphorylated form of RNApII (8WG16) was performed as previously described.10,17,18 ChIP-seq data for runt-related transcription factor 1 (Runx1)19 were obtained from the Gene Expression Omnibus database under accession number SRR364255. ChIP-seq for E26 avian leukemia oncogene 1, 5′ domain (Ets-1), myelocytomatosis oncogene (c-Myc), GATA binding protein 1 (GATA-1), GATA binding protein 2 (GATA-2), and T cell acute lymphocytic leukemia 1 (Tal-1) were obtained from the Encyclopedia of DNA Elements (ENCODE) project (supplemental Table 7).20 Sequence reads were aligned to the mouse genome assembly mm9 with bowtie 0.12.7,21 allowing up to 2 mismatches and keeping only uniquely aligned reads. Tag density tracks and peak calling were generated with MACS 1.4.2.22
Results
Enhancer-associated epigenetic marks at the Ikzf1 lymphoid-specific DHS
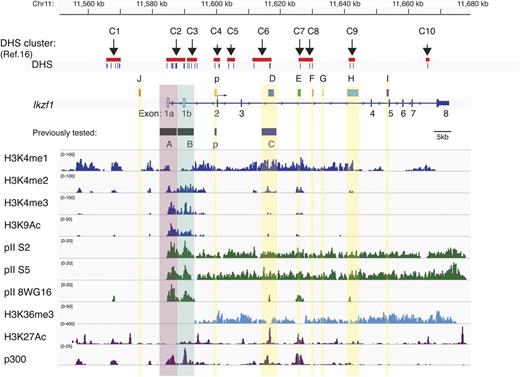
Mapping of lymphoid-specific clusters of DHSs at the Ikzf1 locus has previously identified 10 putative regulatory regions16 (Figure 1). The majority of these sites were also recently identified by genomewide DHS mapping in thymocytes, splenocytes, and B cells generated by the ENCODE project20 (supplemental Figure 1; supplemental Tables 6 and 8). In proximity to these DHS sites were regions of extensive conservation between the mouse and human Ikzf1 (IKZF1). Seven intronic (p, D, E, F, G, H, and I) and 1 upstream (J) conserved regions were identified, and 5 (D-H) were located within the ∼40-kb intron between exons 3 and 4 (>86%; Table 1; Figure 1).
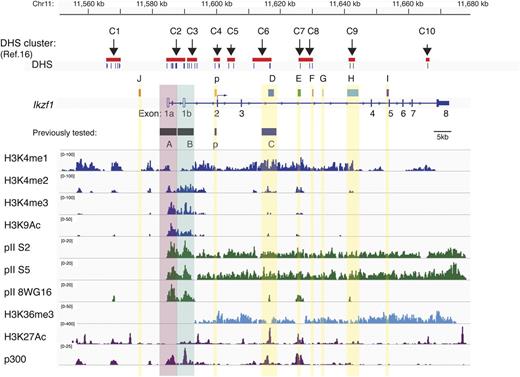
Epigenetic demarcation of Ikzf1 regulatory regions. The location of 10 previously described lymphoid-specific DHS clusters16 and noncoding regions with extensive mouse-human sequence conservation are shown at the Ikzf1 locus. Individual DHS and DHS clusters are indicated by short vertical bars or red bars. On the Ikzf1 locus, the untranslated exons 1a and 1b associated with distinct promoter activities are denoted as unfilled rectangles and the 7 translated exons 2 to 8 as solid rectangles. Enrichment peaks for histone H3 modifications (H3K4me1, me2, me3, H3K9/14Ac, and H3K36me3) and for the RNA Pol II (pII) initiation (8WG16) and elongation (S5, S2) forms at the Ikzf1 locus are displayed by the Integrative Genomics Viewer (IGV) 2.1 using the mouse genome database mm9. Enrichment peaks for H3K27Ac and p300 were deduced from ENCODE data (supplemental Table 8).20
Epigenetic demarcation of Ikzf1 regulatory regions. The location of 10 previously described lymphoid-specific DHS clusters16 and noncoding regions with extensive mouse-human sequence conservation are shown at the Ikzf1 locus. Individual DHS and DHS clusters are indicated by short vertical bars or red bars. On the Ikzf1 locus, the untranslated exons 1a and 1b associated with distinct promoter activities are denoted as unfilled rectangles and the 7 translated exons 2 to 8 as solid rectangles. Enrichment peaks for histone H3 modifications (H3K4me1, me2, me3, H3K9/14Ac, and H3K36me3) and for the RNA Pol II (pII) initiation (8WG16) and elongation (S5, S2) forms at the Ikzf1 locus are displayed by the Integrative Genomics Viewer (IGV) 2.1 using the mouse genome database mm9. Enrichment peaks for H3K27Ac and p300 were deduced from ENCODE data (supplemental Table 8).20
To gain insight into the types of regulatory elements associated with the Ikzf1 conserved DHSs, the immediate chromatin environment was examined by ChIP of histone modifications coupled with high-throughput sequencing (ChIP-Seq) in thymocytes.17 We first examined the methylation status of H3K4 that correlates with both transcriptionally poised and transcriptionally active promoters and enhancers.23 H3K4me1 exhibited a wide distribution over the entire Ikzf1 locus. In contrast, H3K4me2 and H3K4me3 were specifically enriched at the Ikzf1 putative regulatory sites and not throughout the locus (J, p, D, E, F, G, H, and I). Nonetheless, their distributions differed, with H3K4me3 being highly enriched at promoters and relatively depleted at the 5′ and intronic DHS, whereas H3K4me2 was evenly distributed at all these sites. Similar to H3K4me3, H3K9Ac displayed strong promoter enrichment and lower enrichment at other regulatory sites. Similar to H3K4me2, the enhancer marks p300 and H3K27Ac20 showed a nonpromoter distribution.
Transcriptionally active enhancers at the Ikzf1 locus were further evaluated by examining the distribution of the preinitiation and elongation forms of RNApII.24 The S5 and S2 phosphorylated forms of RNA polymerase, as well as the transcriptional elongation mark H3K36me3, were distributed throughout the Ikzf1 locus (Figure 1). In contrast, the unphosphorylated form of RNA polymerase (recognized by the 8WG16 antibody) that marks the preinitiation phase of transcription and active enhancers was only detected at the Ikzf1 regulatory clusters.
Thus, the differential distribution of histone codes, chromatin modifiers, and transcription factors at several of the Ikzf1 conserved DHS clusters supports their potential role as transcriptional enhancers in regulating Ikaros expression during development.
Epigenetic and transcriptional properties of the Ikzf1 enhancers
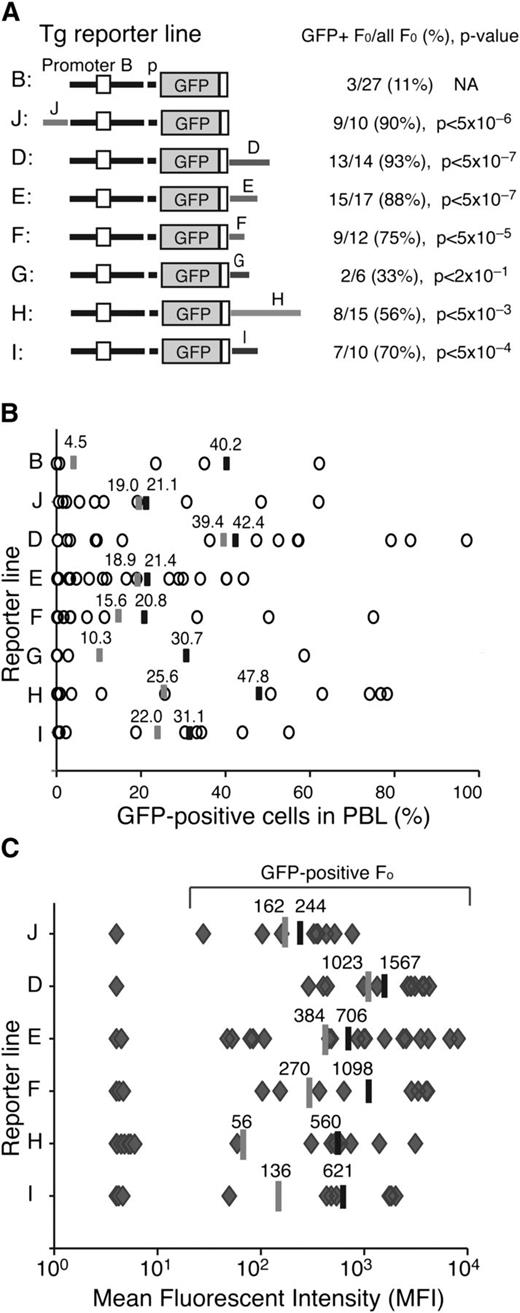
The putative Ikzf1 enhancers were tested for their ability to promote transcription of a GFP reporter from the main Ikzf1 (B-p) promoter (Figures 1 and 2A).16 Reporter cassettes were generated in which the putative enhancer orientation relative to the promoter was preserved, as in the Ikzf1 locus. For each reporter cassette, multiple mouse transgenic lines were generated and tested for GFP expression in peripheral blood leukocytes (PBLs). The number of lines for each reporter and the transgene copy number are provided (supplemental Figures 2 and 3; supplemental Table 1).
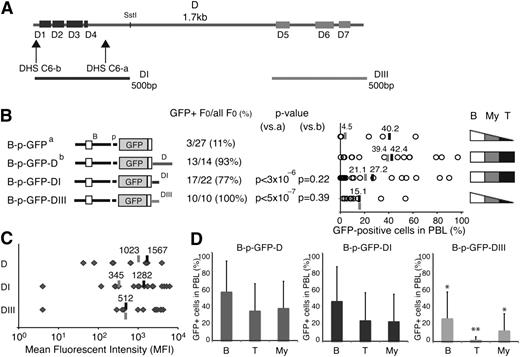
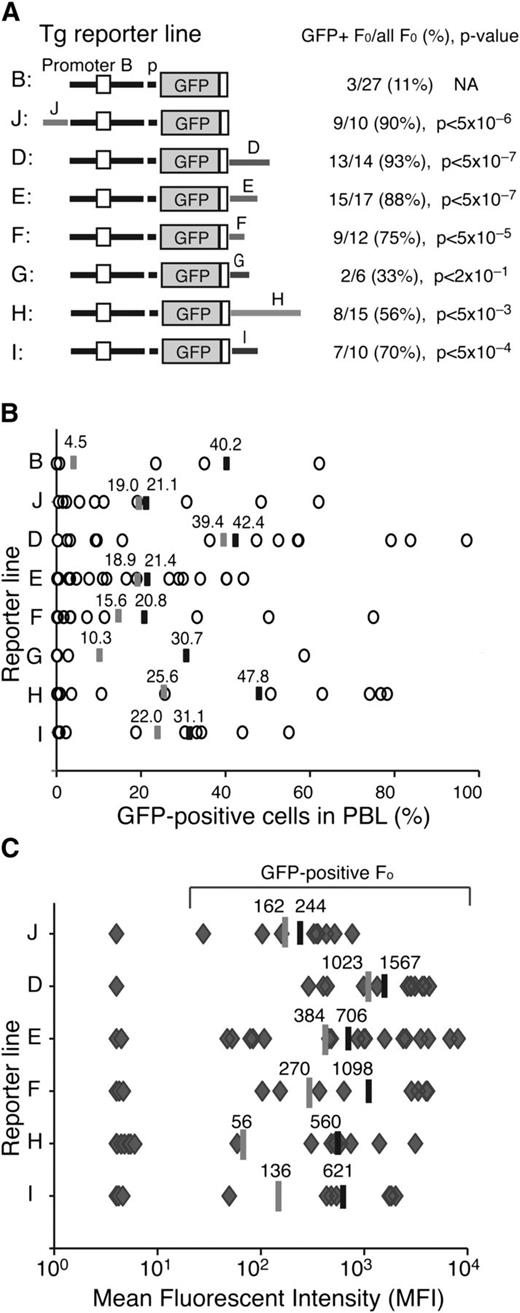
Enhancer activities associated with the Ikzf1 regulatory sites. (A) Diagrams of the reporter constructs used in this study are shown on the left. The number of GFP-expressing founders is displayed over the total number of founders obtained for each reporter. The statistical significance of the difference (increase) in GFP-expressing founder lines compared with the parental B-p-GFP lines was provided by χ2 analysis and is shown as a P value. (B) The percentage of GFP+ PBLs assessed by flow cytometry is depicted for each founder line as circles. The average percentage of GFP+ PBLs for each transgenic reporter was calculated for either all GFP-expressing founders (black bar) or all founders (gray bar). (C) The MFI of GFP+ PBLs was estimated for each founder line (gray diamonds) of every transgenic reporter as described in panel B.
Enhancer activities associated with the Ikzf1 regulatory sites. (A) Diagrams of the reporter constructs used in this study are shown on the left. The number of GFP-expressing founders is displayed over the total number of founders obtained for each reporter. The statistical significance of the difference (increase) in GFP-expressing founder lines compared with the parental B-p-GFP lines was provided by χ2 analysis and is shown as a P value. (B) The percentage of GFP+ PBLs assessed by flow cytometry is depicted for each founder line as circles. The average percentage of GFP+ PBLs for each transgenic reporter was calculated for either all GFP-expressing founders (black bar) or all founders (gray bar). (C) The MFI of GFP+ PBLs was estimated for each founder line (gray diamonds) of every transgenic reporter as described in panel B.
The activity of the Ikzf1 regulatory regions in counteracting local chromatin silencing and in promoting transcription were evaluated by estimating the number of GFP expressing transgenic lines (>1% expression in PBLs), the expression in PBL subsets, and the average level of reporter expression. The majority of transgenic lines generated by these enhancer-driven reporters exhibited GFP expression in PBL (Figure 2A-B). This was a significant increase (P < .05) over the small fraction of GFP-expressing founders obtained with the just-promoter-driven parental reporter (Figure 2A; B-p-GFP, 11%).16 Among the Ikzf1 regulatory regions, J, D, and E displayed the strongest enhancer activity, supporting expression in 88% to 93% of the transgenic lines, whereas F, I, and H were weaker, supporting expression in 53% to 75% of the transgenic lines (Figure 2A; supplemental Figures 2 and 3; supplemental Table 1).
Reporter expression in PBLs was highly variegated (Figure 2B), indicating that although the Ikzf1 enhancers could initially counter-restrictive chromatin at the site of reporter integration, they were unable to maintain transcriptionally permissive chromatin within the Ikzf1-expressing PBL subsets. For example, although the B-p-GFP-D reporter was expressed in 93% of its transgenic lines (P < 5 × 107), on average, expression was detected in only 42% of PBLs. On the other hand, although the B-p-GFP-H reporter was expressed in a smaller fraction (53%) of founder lines, expression was seen in a larger fraction (48%) of PBLs, indicating a stronger potential in maintaining transcriptionally permissive chromatin (Figure 2B).
The Ikzf1 enhancers were further evaluated by measuring the level of reporter expression. Notably, the mean fluorescence intensity (MFI) of GFP-expressing cells from the D enhancer-reporter lines surpassed those of all other reporters, whether measured from all transgenic lines or from expressing transgenic lines (Figure 2C, average shown as gray or black line, respectively). The considerable difference in expression provided by the average MFI of all transgenic vs expressing transgenic lines for F, H, and I suggested that these enhancers were potent in stimulating transcription but only in permissive chromatin. In contrast, although the upstream J enhancer was weak in stimulating transcription, it was very efficient in counteracting repressive chromatin, providing a greater number of low-expressing transgenic lines.
Thus, analysis of transgenic reporters revealed the presence of 2 types of cis-acting elements associated with Ikzf1 enhancers (supplemental Table 1). One type countered restrictive chromatin and position effect variegation (PEV) as documented by an increase in both the number of expressing founders and reporter-expressing PBLs. These elements likely support recruitment of epigenetic factors that promote a transcriptionally permissive chromatin state. The second type was potent in stimulating transcription in permissive chromatin, in line with the activity of a classical enhancer that functions by promoting the initiation or elongation phase of the RNApII cycle.
Cell type specificity of the Ikzf1 enhancers
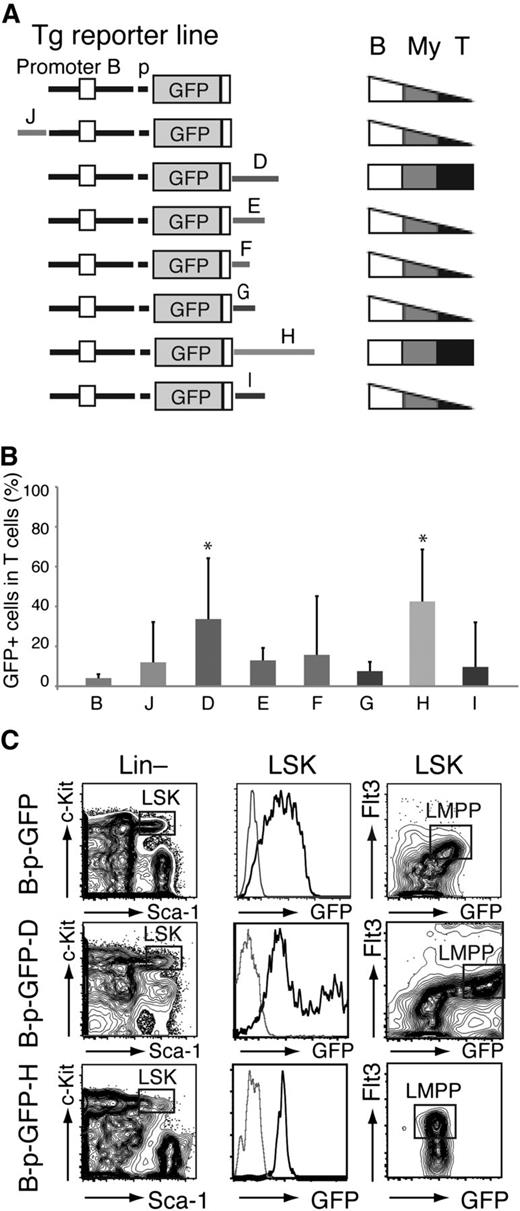
The Ikzf1 enhancers were next evaluated for their cell type–specific effects on reporter expression (Figure 3A-B; supplemental Figures 2, 3, and 5). The expression pattern of their transgenic lines was examined for deviations from the “high B/low T/intermediate myeloid” profile provided by the parental just-promoter-driven reporter on which these enhancers were tested (Figure 3A-B; B-p-GFP).16 PBLs from each founder line were examined for GFP expression in B cells (B220+), T cells (TCRβ+), and myeloid cells (Mac-1+) in peripheral blood by flow cytometry (Figure 3A; supplemental Figures 2, 3, and 5).
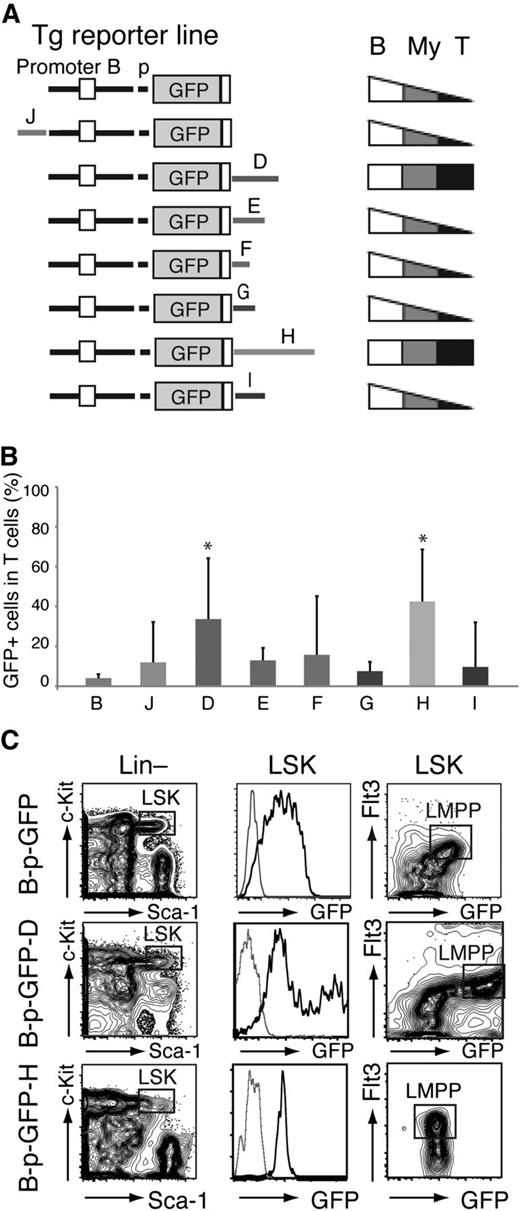
Cell type specificity of the Ikzf1 enhancers. (A) Diagrams of the reporter constructs used in this study are shown on the left. Patterns of lineage-specific GFP expression in PB− B cells (B220+, B), T cells (TCRβ+, T), and myeloid (Mac-1+, My) cells are summarized on the right. (B) Ikzf1 enhancer activity in T cells. Error bars (standard deviation) indicate variegation of GFP expression among the different founder lines generated by each enhancer-based reporter. The significance in the difference of GFP expression among GFP-reporter lines was assessed by Student t test; *P < 5 × 10−3. (C) Enhancer D and H activity in the HSC/MPP-enriched LSK (Lin–c-Kit+Sca-1+), as determined by expression of B-p-GFP-D and -H reporters compared with the parental B-p-GFP reporter. The LMPP is defined as LSK Flt3+ (square gate). GFP expression in the LSK is shown as a histogram or together with Flt3 expression as a contour plot. Thin gray line histogram represents a reporter negative control.
Cell type specificity of the Ikzf1 enhancers. (A) Diagrams of the reporter constructs used in this study are shown on the left. Patterns of lineage-specific GFP expression in PB− B cells (B220+, B), T cells (TCRβ+, T), and myeloid (Mac-1+, My) cells are summarized on the right. (B) Ikzf1 enhancer activity in T cells. Error bars (standard deviation) indicate variegation of GFP expression among the different founder lines generated by each enhancer-based reporter. The significance in the difference of GFP expression among GFP-reporter lines was assessed by Student t test; *P < 5 × 10−3. (C) Enhancer D and H activity in the HSC/MPP-enriched LSK (Lin–c-Kit+Sca-1+), as determined by expression of B-p-GFP-D and -H reporters compared with the parental B-p-GFP reporter. The LMPP is defined as LSK Flt3+ (square gate). GFP expression in the LSK is shown as a histogram or together with Flt3 expression as a contour plot. Thin gray line histogram represents a reporter negative control.
The majority of founder lines generated by the J, E, F, G, and I reporter cassettes displayed a similar expression pattern to the parental cassette (Figure 3A-B; supplemental Figures 2 and 5; supplemental Table 1). In sharp contrast, a significantly large number of the D and H founder lines expressed GFP in a major fraction of T cells compared with the parental reporter (34% and 43% compared with 4%; Figure 3A-B; supplemental Figures 2 and 3; supplemental Table 1). The average proportion of GFP+ myeloid cells was also highest in the D transgenic lines (37% vs 22%; supplemental Figures 2 and 3; supplemental Table 1). Of all the Ikzf1 enhancers tested here, D was the only one capable of stimulating reporter expression in the LMPP, the first step in lymphoid lineage restriction, above the basal levels detected in the HSC-enriched population (Figure 3C, LSK Flt3+ vs LSK Flt3–).4
Thus, whereas all 6 of the Ikzf1 enhancers (J, D, E, F, H, and I) were active in B and myeloid cells, 2 (D and H) were also active in T cells and were likely responsible for high expression of Ikzf1 in thymocytes required for normal maturation and homeostasis.1,9,17,25,26 One of the 2 Ikzf1 T cell–specific enhancers (D) was active in the LMPP and was associated with both an increase in Ikaros expression and induction of lymphoid lineage differentiation potential.4,6
Enhancer D plays a nonredundant role in the transcriptional regulation of Ikzf1
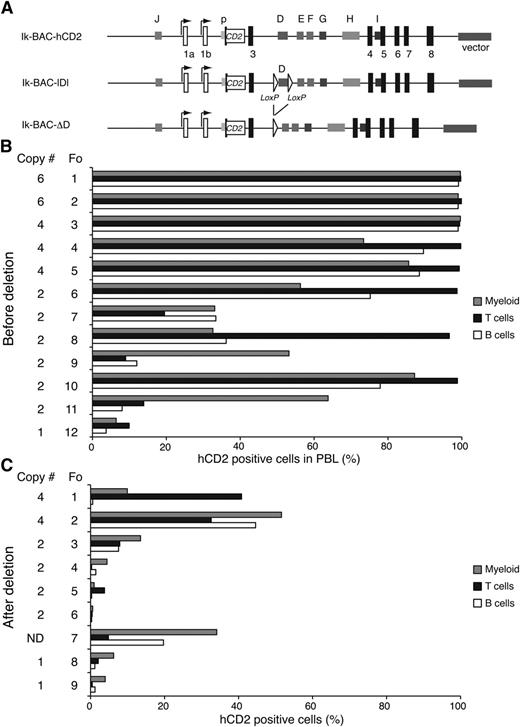
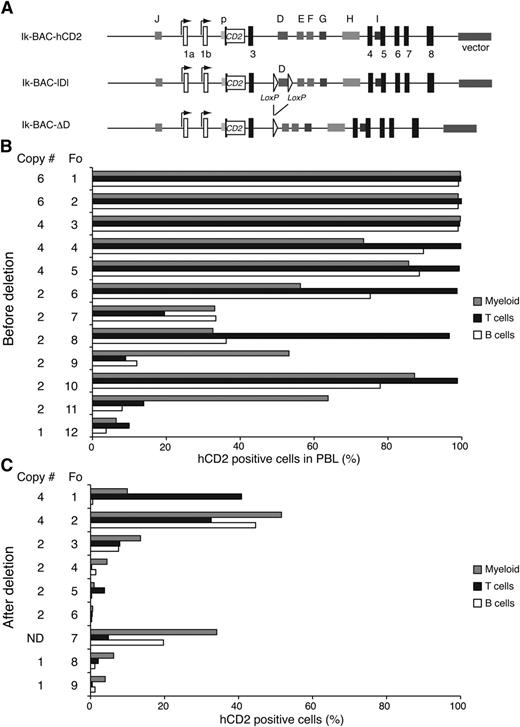
Given the potential importance of the enhancer D, we examined how its deletion affected transcriptional activity at the Ikzf1 locus. An Ikzf1 BAC clone with the human CD2 (hCD2) reporter inserted in exon 2 (Ik-BAC-hCD2) was engineered to contain loxP recognition sites flanking the 1.7-kb D region and was tested for expression in lymphoid and myeloid cells prior to (Ik-BAC-lDl) and after Cre-mediated deletion (Ik-BAC-ΔD) (Figure 4A). Because these BAC vectors were injected into fertilized eggs in a circular form, we presumed that transgenic lines with ≥3 copies of the transgene had ≥1 intact BAC clone. All of the founders obtained with Ik-BAC-hCD2 and Ik-BAC-lDl were expressed in ∼90% of T, B, and myeloid cells, even when 2 copies of the BAC transgene were present (Figure 4B). In contrast, in all of the founders obtained with Ik-BAC-ΔD, a highly variegated expression of the hCD2 reporter was detected in all cell types, irrespective of the copy number of the transgene (Figure 4C). Thus, enhancer D is critical in counteracting repressive chromatin effects at the Ikzf1 locus and maintaining transcription at high levels, an activity that cannot be substituted by the other Ikzf1 enhancers.
Enhancer D is required for normal transcription at the Ikzf1 locus (A) Diagram of the Ikzf1 BAC transgenic reporter constructs. The first translated exon (exon 2) in the Ikzf1 BAC clone was replaced by the human CD2 (hCD2) reporter gene (white rectangle) inserted at the initiation codon (Ik-BAC-CD2). Two flanking loxP sites (white triangles) were inserted 5′ and 3′ of the D region (Ik-BAC-lDl). The D region was deleted from Ik-BAC-lDl by Cre recombinase to generate Ik-BAC-ΔD construct. (B) Reporter expression in the PBL from Ik-BAC transgenic lines with intact enhancer D region. (C) Reporter expression in PBL from Ik-BAC-ΔD transgenic lines. The copy number is noted besides each founder generated from either the Ik-BAC-CD2 or the Ik-BAC-DL line. The percentage of hCD2+ myeloid cells (gray bars), T cells (black bars), and B cells (white bars) was determined for each founder animal by flow cytometry with antibodies to Mac-1, B220, and TCRβ, respectively. N.D., not determined.
Enhancer D is required for normal transcription at the Ikzf1 locus (A) Diagram of the Ikzf1 BAC transgenic reporter constructs. The first translated exon (exon 2) in the Ikzf1 BAC clone was replaced by the human CD2 (hCD2) reporter gene (white rectangle) inserted at the initiation codon (Ik-BAC-CD2). Two flanking loxP sites (white triangles) were inserted 5′ and 3′ of the D region (Ik-BAC-lDl). The D region was deleted from Ik-BAC-lDl by Cre recombinase to generate Ik-BAC-ΔD construct. (B) Reporter expression in the PBL from Ik-BAC transgenic lines with intact enhancer D region. (C) Reporter expression in PBL from Ik-BAC-ΔD transgenic lines. The copy number is noted besides each founder generated from either the Ik-BAC-CD2 or the Ik-BAC-DL line. The percentage of hCD2+ myeloid cells (gray bars), T cells (black bars), and B cells (white bars) was determined for each founder animal by flow cytometry with antibodies to Mac-1, B220, and TCRβ, respectively. N.D., not determined.
Characterization of the cis-acting elements of the Ikzf1 enhancer D
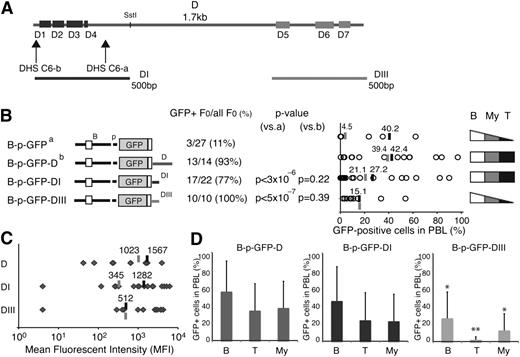
We further characterized the cis-acting elements responsible for enhancer D activities. The 1.7-kb region encompassing this enhancer contained 7 smaller areas (D1-D7) of strong mouse-human conservation (86%-96%) that ranged in size from 37 to 103 bp (Figure 5A; supplemental Figure 6; supplemental Table 3). The first 4 areas (D1-D4) were clustered at the 5′ end and were associated with the 2 DHS-C6 sites (Figure 5A; DHS-C6-a and DHS-C6-b), whereas the other 3 sites (D5-D7) covered a total of 394 bases located at the 3′ end (Figure 5A). Two 500-bp fragments encompassing the D1 to D4 (DI) and D5 to D7 (DIII) sequences, respectively, were cloned into the B-p-GFP cassette and evaluated for activity compared with the 2 parental vectors (Figure 5B; B-p-GFP and B-p-GFP-D).
T cell–specific cis-acting elements in enhancer D. (A) Areas of sequence conservation (D1-D7) within the 1.7-kb enhancer D are shown as rectangles. Horizontal lines below the D region depict the two 500-bp DI and DIII subregions used in transgenic reporters. (B) Comparison of reporter expression between the B-p-GFP-DI, B-p-GFP-DIII, B-p-GFP, and B-p-GFP-D founder lines. The reporter constructs used to generate these founder lines are depicted on the left. The number of GFP-expressing founders is displayed over the total number of founders generated. The statistical significance of the difference (increase or decrease) in the proportion of GFP-expressing founder compared with the parental B-p-GFP line (a) or B-p-GFP-D (b) was provided by χ2 analysis and is shown as a P value. The percentage of GFP-positive PBLs is depicted for each founder line (circle). The average percentage of GFP+ PBLs for each transgenic reporter was calculated for either all GFP-expressing founders (black bar) or all founders (gray bar). The cell type specificity of each reporter is depicted on the right. (C) The MFI of GFP+ PBLs from B-p-GFP-D, B-p-GFP-D-I, and B-p-GFP-D-III transgenic lines was calculated for each founder (gray diamonds). Gray lines indicate the average MFI among all founders, and black lines denote the average MFI among the GFP-expressing founders. (D) Lineage-specific GFP expression in the PBLs from B-p-GFP-D, B-p-GFP-DI, and B-p-GFP-DIII founders. The average percentage of GFP+ cells within peripheral blood B cells (B), T cells (T), and myeloid cells (My) is shown. The significance in the difference of expression among GFP reporters was assessed by Student t test; *P < 5 × 10−2; **P < 5 × 10−3. Error bars (standard deviation) indicate variegation of GFP expression among the different founder lines made by each enhancer-based reporter.
T cell–specific cis-acting elements in enhancer D. (A) Areas of sequence conservation (D1-D7) within the 1.7-kb enhancer D are shown as rectangles. Horizontal lines below the D region depict the two 500-bp DI and DIII subregions used in transgenic reporters. (B) Comparison of reporter expression between the B-p-GFP-DI, B-p-GFP-DIII, B-p-GFP, and B-p-GFP-D founder lines. The reporter constructs used to generate these founder lines are depicted on the left. The number of GFP-expressing founders is displayed over the total number of founders generated. The statistical significance of the difference (increase or decrease) in the proportion of GFP-expressing founder compared with the parental B-p-GFP line (a) or B-p-GFP-D (b) was provided by χ2 analysis and is shown as a P value. The percentage of GFP-positive PBLs is depicted for each founder line (circle). The average percentage of GFP+ PBLs for each transgenic reporter was calculated for either all GFP-expressing founders (black bar) or all founders (gray bar). The cell type specificity of each reporter is depicted on the right. (C) The MFI of GFP+ PBLs from B-p-GFP-D, B-p-GFP-D-I, and B-p-GFP-D-III transgenic lines was calculated for each founder (gray diamonds). Gray lines indicate the average MFI among all founders, and black lines denote the average MFI among the GFP-expressing founders. (D) Lineage-specific GFP expression in the PBLs from B-p-GFP-D, B-p-GFP-DI, and B-p-GFP-DIII founders. The average percentage of GFP+ cells within peripheral blood B cells (B), T cells (T), and myeloid cells (My) is shown. The significance in the difference of expression among GFP reporters was assessed by Student t test; *P < 5 × 10−2; **P < 5 × 10−3. Error bars (standard deviation) indicate variegation of GFP expression among the different founder lines made by each enhancer-based reporter.
Both enhancer subdomains generated a high number of expressing founder lines that were comparable to that of the intact enhancer D (Figure 5B). However, within the expressing populations, reporter variegation was greater compared with that of the intact enhancer. On average, 42% of PBLs from the D founder lines expressed GFP but only 27% or 15% of PBL from the DI or DIII founders expressed GFP, respectively (Figure 5B; supplemental Table 2).
The level of reporter expression supported by the 2 enhancer D subdomains differed greatly, those by DIII being much lower compared with DI or with the intact parental enhancer (Figure 5C-D; MFI: 512 vs 1282 or 512 vs 1567). Expression supported by DI was comparable to that of the intact enhancer (Figure 5C; MFI: 1282 vs 1567). Furthermore, DI but not DIII was able to confer expression in a substantial fraction of T cells (Figure 5B,D; supplemental Figure 4; supplemental Table 2). By these criteria, the DI subregion had similar transcriptional and cell type–specific properties as the full-length enhancer D.
Combination of Ikzf1 enhancers functions as a locus control region
When tested individually, the Ikzf1 enhancers were capable of appreciable but not complete alleviation of PEV on reporter expression (Figure 2). This suggests that the combined enhancer activities, as encountered in the Ikzf1 locus, are required for proper Ikaros expression in lympho-myeloid cells.
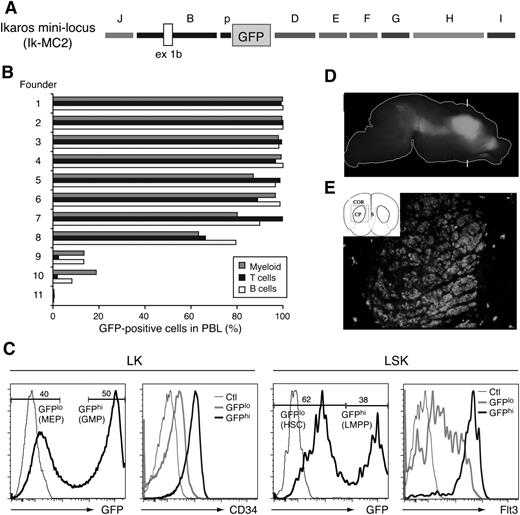
To test this hypothesis, we generated a transgenic reporter that combined most of these elements, preserving their orientation as in the endogenous locus (Ikaros mini-regulatory locus [Ik-MC2]; Figure 6A). Eight of 11 transgene positive founders exhibited GFP reporter expression in almost 100% of B cells, T cells, and myeloid cells in PBLs (Figure 6B). The 3 founders with low reporter expression may be due to integration of an incomplete transgenic reporter in the genome. Thus, the combined activity of regulatory elements present in the Ik-MC2 cassette can counteract PEV and provide an endogenous Ikaros-like pattern of expression in PBLs (Figure 6B).
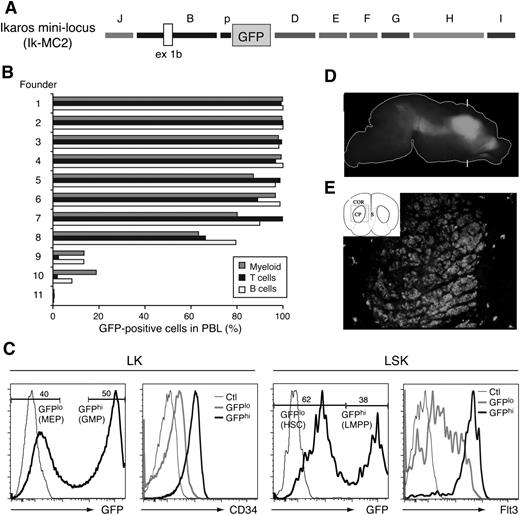
Combined activity of Ikzf1 regulatory elements in lympho-myeloid and neuronal cells. (A) Diagram of the Ik-MC2 construct. (B) GFP reporter expression in PBL subsets of the Ik-MC2 founder lines. The percentage of GFP+ myeloid cells (gray bars), T cells (black bars), and B cells (white bars) was determined for each founder line by fluorescence-activated cell sorter (FACS) analysis. (C) GFP expression in the HSC-enriched (LSK) and erythro-myeloid (LK-Lin–c-Kit+Sca-1–) progenitor populations of the bone marrow. CD34 and Flt3 expression in LSK and LK GFP high (hi) and low (lo) subsets are shown. Thin line histogram represents FACS staining of a reporter or marker negative control. (D) Lateral view of GFP expression in the P0 brain in whole mount. The outline of the brain is shown as a white line. Bright GFP expression is observed in the basal ganglia. No fluorescence was observed in wild-type controls under these conditions. Brackets mark the approximate plane of the frontal section shown in panel E. (E) Schematic view of the section indicating the cortex (COR), caudate putamen (CP), and septum (S) and the area shown in the photograph (box). GFP expression is observed in the caudate putamen. With the exception of scattered cells elsewhere, GFP is not observed in surrounding tissue.
Combined activity of Ikzf1 regulatory elements in lympho-myeloid and neuronal cells. (A) Diagram of the Ik-MC2 construct. (B) GFP reporter expression in PBL subsets of the Ik-MC2 founder lines. The percentage of GFP+ myeloid cells (gray bars), T cells (black bars), and B cells (white bars) was determined for each founder line by fluorescence-activated cell sorter (FACS) analysis. (C) GFP expression in the HSC-enriched (LSK) and erythro-myeloid (LK-Lin–c-Kit+Sca-1–) progenitor populations of the bone marrow. CD34 and Flt3 expression in LSK and LK GFP high (hi) and low (lo) subsets are shown. Thin line histogram represents FACS staining of a reporter or marker negative control. (D) Lateral view of GFP expression in the P0 brain in whole mount. The outline of the brain is shown as a white line. Bright GFP expression is observed in the basal ganglia. No fluorescence was observed in wild-type controls under these conditions. Brackets mark the approximate plane of the frontal section shown in panel E. (E) Schematic view of the section indicating the cortex (COR), caudate putamen (CP), and septum (S) and the area shown in the photograph (box). GFP expression is observed in the caudate putamen. With the exception of scattered cells elsewhere, GFP is not observed in surrounding tissue.
We also evaluated the expression profile of the Ik-MC2 cassette in the HSCs and in early lineage restricted progenitors. This was very similar to that previously described for the enhancer D–containing reporters (Figure 3C; B-p-GFP-D and B-p-GFP-C).4 The bimodal expression of the GFP reporter supported by the Ik-MC2 cassette separated the LMPP from the multipotent HSC (Figure 6C; LSK Flt3++: GFPhi from LSK Flt3neg–lo: GFPlo). It also segregated granulocyte-macrophage progenitors from megakaryocyte-erythrocyte progenitors in a mixed erythro-myeloid progenitor population (Figure 6C; LK CD34+: GFPhi from LK CD34neg-lo: GFP+).4
Ikzf1 is also expressed is in the developing striatum,27,28 where it may play a role in neural progenitor function.29,30 In contrast to enhancer D or other Ikzf1 enhancers tested in isolation, their combined activity supported GFP expression in the striatum (Figure 6D-E). Sections through this region of the brain revealed expression throughout the caudate putamen similar to that exhibited by the endogenous Ikzf1 locus (Figure 6D-E).
Thus, the combination of 9 of 10 of the Ikzf1 conserved regions (J, B, p, D, E, F, G, H, and I) in a miniregulatory locus ensured gene expression in cells of the lympho-myeloid and neuronal lineage in a manner that resembled the cell type specificity of the endogenous Ikzf1 locus and that was unaffected by local chromatin silencing effects.
Network of transcription factors regulating Ikzf1 expression
To obtain insight into the transcription factors involved in Ikzf1 regulation, motif search for transcription factor binding sites at enhancers D and H was performed. Enhancer D was specifically enriched with binding sites for transcription factors expressed in early hematopoietic progenitors such as Runx (Runx1-3), Homeobox A9 (Hoxa9), special AT-rich sequence binding protein 1 (Satb1), interferon regulatory factors (Irf1, Irf4), CCAAT/enhancer binding proteins (Cebpa, Cepbb), and myocyte enhancer factor 2C (Mef2c) (Figure 7B; supplemental Table 7D-H). These early lympho-myeloid transcription factor binding sites were not present in enhancer H. Nonetheless, several shared factor binding sites for transcription factors with previously reported activity in T cells were present in enhancers D and H, such as E2A (Tcf3), Ets-1 (Ets1), Tal-1 (Tal1), and Ikaros (Ikzf1) (supplemental Table 7D,H; Figure 7B). Further dissection of D into the DI and DIII subregions provided independent evidence that the progenitor and T cell–specific activities of enhancer D resided within the DI subregion (supplemental Table 7DI-DIII; Figure 7B).
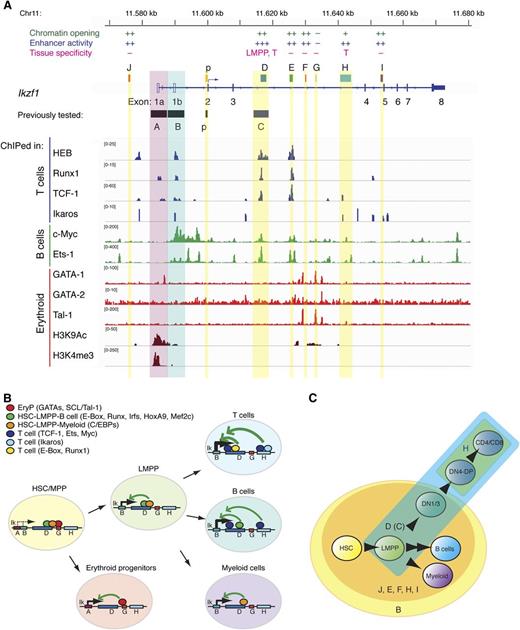
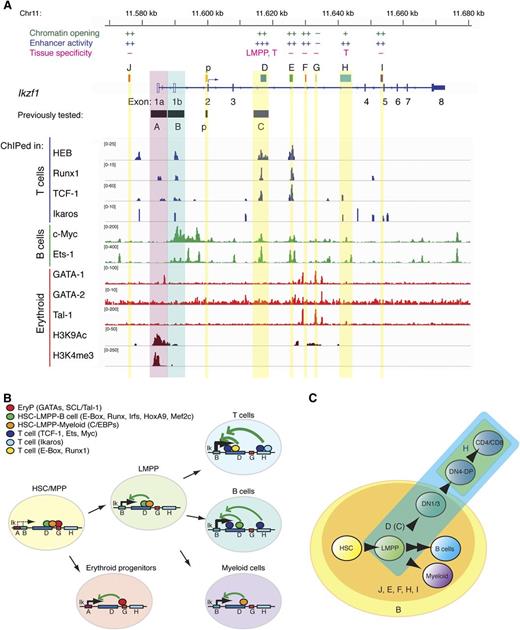
Network of transcription factors targeting Ikzf1 enhancers. (A) TF enrichment peaks at the Ikzf1 locus in thymocytes (T cells), CH12 (B cells), and erythoid precursors were visualized by the IGV browser. A summary of regulatory activities is provided above respective enhancer regions at the Ikzf1 locus. (B-C) A model of Ikzf1 regulation. (B) Ikzf1 enhancer-promoter interactions during hematopoiesis. Erythroid-specific factors binding at enhancer G enable interactions with the myeloid-specific promoter A and Ikaros expression during early erythropoiesis. Lympho-myeloid–specific factors binding at enhancers D and H support interactions with the lympho-myeloid–specific promoter B and Ikaros expression during lymphoid and myeloid differentiation. Lineage-specific transcription factors at these sites are depicted as color-coded circles. Arrows indicate potential interactions between cell type–specific enhancers and promoters supporting Ikzf1 expression at appropriate developmental stages. (C) Ikzf1 regulatory region activity during lymphopoiesis. The Ikzf1 lympho-myeloid promoter B, although active from the HSCs through B cell and myeloid differentiation, requires input from an enhancer to overcome the restrictive chromatin effects and PEV (yellow circle). Enhancers J, D(C), E, F, H, and I counteract PEV and raise Ikzf1 gene expression (orange circle). For Ikzf1 expression past the DN stage of T-cell development, input is required from enhancers D(C) (blue) and H (green). Induction of Ikzf1 expression in the LMPP to the level required for lymphocyte differentiation is dependent on enhancer D(C) activity (blue).
Network of transcription factors targeting Ikzf1 enhancers. (A) TF enrichment peaks at the Ikzf1 locus in thymocytes (T cells), CH12 (B cells), and erythoid precursors were visualized by the IGV browser. A summary of regulatory activities is provided above respective enhancer regions at the Ikzf1 locus. (B-C) A model of Ikzf1 regulation. (B) Ikzf1 enhancer-promoter interactions during hematopoiesis. Erythroid-specific factors binding at enhancer G enable interactions with the myeloid-specific promoter A and Ikaros expression during early erythropoiesis. Lympho-myeloid–specific factors binding at enhancers D and H support interactions with the lympho-myeloid–specific promoter B and Ikaros expression during lymphoid and myeloid differentiation. Lineage-specific transcription factors at these sites are depicted as color-coded circles. Arrows indicate potential interactions between cell type–specific enhancers and promoters supporting Ikzf1 expression at appropriate developmental stages. (C) Ikzf1 regulatory region activity during lymphopoiesis. The Ikzf1 lympho-myeloid promoter B, although active from the HSCs through B cell and myeloid differentiation, requires input from an enhancer to overcome the restrictive chromatin effects and PEV (yellow circle). Enhancers J, D(C), E, F, H, and I counteract PEV and raise Ikzf1 gene expression (orange circle). For Ikzf1 expression past the DN stage of T-cell development, input is required from enhancers D(C) (blue) and H (green). Induction of Ikzf1 expression in the LMPP to the level required for lymphocyte differentiation is dependent on enhancer D(C) activity (blue).
Transcription factors with putative binding sites in the Ikzf1 regulatory elements were further evaluated by analysis of ChIP-seq data at the Ikzf1 locus17,19,31 (Figure 7A). Many of the lymphoid lineage pioneering factors such as Ikaros, HEB, Runx1, and TCF-1 bound at multiple sites in the Ikzf1 locus. Enhancer D supported strong enrichment of a multitude of transcription factors expressed from the early to the later stages of lymphoid differentiation (E-box/HEB, Runx1, TCF-1, Ets-1, c-Myc), consistent with the enhancer’s broad activity from the early progenitors to later stages in lymphocyte differentiation and its key role in the Ikzf1 regulation. Interestingly, the neighboring enhancer E displayed a similar to D enrichment of transcription factors, although its in vivo activity was more restricted. The H enhancer with activity only in T cells had a limited repertoire of transcription factors that included Ikaros, TCF-1, Ets-1, and c-Myc. Finally, all the lymphoid promoting transcription factors bound to the lympho-myeloid promoter B, whereas binding of the erythroid specific transcription factor GATA-1 was detected in the vicinity of the myeloid-specific promoter A, which was specifically marked by H3K4me3 and H3K9Ac in erythroid progenitors. Analysis of erythroid transcription factor such as GATA-1, GATA-2, and SCL/Tal-1 (Tal1) identified enrichment at F and G regions in the vicinity of H3K9Ac, suggesting that these may be enhancers responsible for supporting Ikaros expression in erythroid progenitors. (Figure 7A-B).
Discussion
The transcriptional regulation of the Ikzf1 gene is key to our understanding of the mechanisms that control normal lymphocyte differentiation and its aberrant manifestations. Here we show that Ikzf1 expression in the hematopoietic system is controlled by a network of cis-acting elements that is spread over a 120-kb genomic region with many shared but also unique epigenetic and transcriptional regulatory features.
A comparative study of previously established maps of lymphoid-specific DHS, with regions of human-mouse sequence conservation at the Ikzf1 locus, identified several cis-acting regulatory elements that are likely responsible for modulating Ikaros expression in this developmental system. Mapping of histone modification signatures and the preinitiation and elongation forms of RNApII at the Ikzf1 locus in thymocytes has provided further insight into the location of promoter and enhancer elements, residing in the vicinity of conserved lymphoid-specific DHS clusters.23,24,32-34 The promoter-demarcating H3K4me3 and H3K9Ac were highly enriched over the previously characterized Ikzf1 lympho-myeloid promoter B. On the other hand, a restricted enrichment of H3K4me2, H3K27Ac, and the preinitiation form of RNApII over the intronic Ikzf1 DHS clusters was consistent with previous reports of their marking active and poised enhancers.23,24,32-34
Because the ultimate proof of enhancer activity is the ability to stimulate expression from the gene’s promoter, the ability of putative enhancers was tested on the Ikzf1 lympho-myeloid promoter in vivo. The Ikzf1 enhancer regions were evaluated for their ability to counteract transcriptionally repressive chromatin, to increase transcriptional rate, and to promote cell type–specific expression of a transgenic reporter during lymphoid and myeloid lineage differentiation. Six of the 7 Ikzf1 conserved regions showed enhancer activities in B cells and myeloid cells (Figure 7A,C). However, only when these enhancers were combined into 1 regulatory unit could they function as an locus control region (LCR) and counteract all restrictive chromatin effects. A similar combinatorial action of distantly located regulatory elements functioning together as an LCR has been described for the λ5 and the V-preB.35
Outside the hemo-lymphoid system, Ikzf1 is expressed in neural precursors of the cortex. In isolation, none of the Ikzf1enhancers were capable of brain-specific reporter expression, but when combined into a single regulatory unit with LCR activity, expression in the appropriate brain cortical region was detected.
Ikzf1 is expressed at high levels in differentiating thymocytes, where it is required for their normal maturation. Loss of Ikaros in thymocytes causes their aberrant expansion and transformation.9,10,17,25,26 The previously identified Ikzf1 promoter is active only in B cells and myeloid cells, indicating that additional transcriptional factor input from an enhancer is needed for T cell–specific expression. Two of the 7 enhancers (D and H) were capable of supporting expression in T cells. The rest of the Ikzf1 enhancers did not confer additional cell type specificity to the Ikzf1 promoter, although they greatly increased promoter activity in B cells and myeloid cells (Figure 7A,C).
In the developing embryo, Ikzf1 expression is first detected in the hemangioblast region and in HSC-enriched compartments, albeit at lower levels compared with fetal sites harboring committed lymphocyte precursors.27 During transition from an MPP to an LMPP,3-5 Ikzf1 expression is upregulated. The LMPP is the first restriction point downstream of the HSC, where upregulation of lymphoid-specific genes and downregulation of stem cell– and erythroid- but not myeloid-specific genes is occurring in an Ikaros-dependent fashion.4,6 Notably, the Ikzf1 enhancer D was the only Ikzf1 regulatory region responsible for augmenting reporter expression in the LMPP (Figure 7B), suggesting that transcriptional inputs from its elements are key to the increase in Ikaros expression and lymphoid lineage specification.4,6
The importance of the Ikzf1 enhancer D was further established in the context of the Ikzf1 locus by its deletion from an Ikzf1 BAC reporter. The enhancer D–deleted BAC reporter exhibited a highly variegated expression compared with the parental reporter. Two highly conserved sequence clusters were mapped at the 5′ and 3′ end of enhancer D (DI and DIII, respectively). Notably, the LMPP and T-cell specificity of enhancer D was ascribed to DI, whereas both clusters supported permissive chromatin.
Insight into the transcriptional networks responsible for inducing Ikzf1 expression at the HSC and the LMPP and during T-cell differentiation was obtained by motif analyses for transcription factor binding sites at key Ikzf1 enhancers and chromatin enrichment of putative transcriptional regulators at the Ikzf1 locus. Both enhancers D and H contained sites for transcription factors highly expressed in T- and B-cell precursors, such as TCF-1, Runx1, HEB (E-Box), Ikaros, Ets1, and c-Myc, that play a determining role in lymphocyte differentiation. Sites for factors expressed early in lymphocyte development, such as HoxA9, E2A, Satb1, and Mef2c, were present only at enhancer D, underscoring its unique activation and function among Ikzf1 enhancers at the onset of lymphopoiesis (Figure 7B-C). Further studies on these regulators with respect to their unique or redundant contribution to Ikzf1 regulation will further establish their importance and position in the regulatory network that underscores lymphoid lineage differentiation.
Inactivating deletions and mutations in the IKZF1 locus have been reported as poor prognostic indicators in pre–B-cell progenitor acute lymphoblastic leukemia and early T-cell progenitor acute lymphoblastic leukemia in humans and mice.12,13,15,36 Large deletions at the 7p arm resulting in the loss of the whole IKZF1 allele, as well as smaller deletions between exons 4 to 7 that produce dominant-negative IKAROS protein isoforms or between exons 2 to 7 that have a potential for generating either a null or dominant negative allele, have been reported. Finally, point mutations that abolish the DNA binding activity of IKAROS and also generate dominant negative IKAROS protein isoforms have been reported in association with acute lymphoblastic leukemias.13,37-39 Mutations in the IKZF1 locus occurring in the highly conserved Ikzf1 regulatory regions may interfere with Ikaros expression and provide a mechanism of leukemia development.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr M. Busslinger for kindly providing IkAB/pQS1, Dr N. Copeland for sharing EL250 cells, Dr H. Kawamoto for sharing the antibody against TCF-1, and Bob Czyzewski for mouse husbandry.
This work was supported by National Institutes of Health Research Project grants 5R37 R01 AI33062, (National Institute of Allergy and Infectious Diseases) and 9R01CA162092-19 (National Cancer Institute) (to K.G.), R21AI076720 (to F.G.), and The Lady Tata Memorial Trust (M.D.).
Authorship
Contribution: T.Y., E.L., M.D., I.H., T.N., A.F.J., J.W., E.A.P., C.K., and B.A.M. performed experiments and analyzed results; J.Z., M.D., T.Y., and F.G. performed bioinformatics analysis; and T.Y., E.L., and K.G. designed the research, made the figures, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for M.D. is National Institutes of Health Genomics & Immunity Section, NIAMS BG10, 13C101a 10 Center Dr, MSC 1930, Bethesda, MD 20892.
The current affiliation for J.Z. is School of Biological Sciences, Room 2S-04, 2/F, Kadoorie Biological Sciences Building, University of Hong Kong, Hong Kong SAR, China.
The current affiliation for T.N. is Department of Immunology, Toho University School of Medicine, Ohta-ku, Tokyo, Japan.
Correspondence: Katia Georgopoulos, Cutaneous Biology Research Center, Massachusetts General Hospital, Harvard Medical School, 149 East 13th St, Charlestown, MA 02129; e-mail: katia.georgopoulos@cbrc2.mgh.harvard.edu; and Toshimi Yoshida, Cutaneous Biology Research Center, Massachusetts General Hospital, Harvard Medical School, 149 East 13th St, Charlestown, MA 02129; e-mail: toshimi.yoshida@cbrc2.mgh.harvard.edu.
References
Author notes
T.Y. and E.L. contributed equally to this work.