Key Points
miR-155 knockdown in myeloid cells accelerates spontaneous breast cancer development.
miR-155 is required by TAMs for deploying antitumoral activity.
Abstract
A productive immune response requires transient upregulation of the microRNA miR-155 in hematopoietic cells mediating innate and adaptive immunity. In order to investigate miR-155 in the context of tumor-associated immune responses, we stably knocked down (KD) miR-155 in the myeloid compartment of MMTV-PyMT mice, a mouse model of spontaneous breast carcinogenesis that closely mimics tumor-host interactions seen in humans. Notably, miR-155/KD significantly accelerated tumor growth by impairing classic activation of tumor-associated macrophages (TAMs). This created an imbalance toward a protumoral microenvironment as evidenced by a lower proportion of CD11c+ TAMs, reduced expression of activation markers, and the skewing of immune cells within the tumor toward an macrophage type 2/T helper 2 response. This study highlights the importance of tumor-infiltrating hematopoietic cells in constraining carcinogenesis and establishes an antitumoral function of a prototypical oncomiR.
Introduction
Innate and adaptive immune responses, modulated and potentially subverted by tumor cells, create an environment of chronic inflammation that can turn a host defense mechanism into a promoter of tumor growth.1 Tumor progression has been associated with the expansion of the myeloid compartment, and myeloid subsets composed of incompletely differentiated cells, often termed “myeloid derived suppressor cells,” have been found to possess protumoral properties in experimental models.2 Within the tumor, myeloid cells accumulate mostly in the form of tumor-associated macrophages (TAMs), and their quantity correlates with worse prognosis in many types of tumors including those from Hodgkin lymphoma and breast cancer.3,4 Instead, TAM infiltration can also be indicative of an improved prognosis, for example, in colorectal cancer.5 TAMs are heterogeneous, with some subsets likely acting as bona fide ag-presenting cells (APCs) eliciting antitumor immunity, and others fulfilling tissue remodeling functions and thus favoring tumor progression by supporting angiogenesis, invasion, and metastasis.6,7 Our group has distinguished these functionally different subsets by the expression of the tyrosine kinase receptor TIE2.6,8 Furthermore, we have identified 2 distinct gene expression signatures in TIE2+ TAMs (TEMs) and TIE2− TAMs, enabling their prospective isolation based on MRC1 and CD11c expression, respectively.9 Whereas these data highlight the heterogeneity of TAMs, little is known on the molecular mechanisms underlying their activation and function within the tumor microenvironment.
MicroRNAs (miRNAs) have emerged as global regulators of gene expression programs.10 Recently, miR-511-3p, an miRNA encoded by the Mrc1 gene and thus associated with the macrophage subset with tissue remodeling function, has been shown to provide a negative feedback on protumoral genetic programs.11 On the other hand, miRNAs can promote inflammation. miR-155 is upregulated by immune stimuli such as antigens, Toll-like receptor ligands, and inflammatory cytokines in cells of the innate and adaptive immune system.12-17 miR-155−/− mice have global immune defects characterized by decreased dendritic cell function and defective B- and T-cell responses.13-15 Specifically, the failure of miR-155−/− mice to acquire protective immunity against Salmonella typhimurium infection was attributed to a failure of dendritic cells to efficiently activate T cells, probably due to a deficiency in antigen presentation and costimulation.13 The adaptive immune response in these mice was skewed toward a T helper 2 (Th2)–type immune response associated with the production of Th2 cytokines including interleukin (IL)-4, IL-5, and IL-10. These data strongly suggest that miR-155 is required for a T helper (Th1)–type response.
In this work, we investigated the function of miR-155 in TAMs using a spontaneous breast cancer mouse model. We found that miR-155 is upregulated in CD11c+ pro-inflammatory TAMs. Using a myeloid cell specific loss of function approach, we established that miR-155 is required for the activation of CD11c+ TAMs, and that this TAM subset, in turn, actively mediates antitumor immunity, especially during the early stages of breast carcinogenesis.
Material and methods
Lentiviral vectors
Cell lines
293T and RAW264.7 cells were maintained in Iscove modified Dulbecco medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum (Gibco) and a combination of penicillin-streptomycin and glutamine.
Mice
FVB and C57BL/6N wild-type mouse strains were purchased from Charles River Laboratories, while FVB/PyMT mice were purchased from the NCI-Frederick Mouse Repositories. All mice were maintained in specific-pathogen-free conditions, and all procedures were performed according to protocols approved by the Animal Care and Use Committee of the Fondazione San Raffaele del Monte Tabor (IACUC 447, 455) and communicated to the Ministry of Health and local authorities according to Italian law.
HSPC transduction and transplantation
Bone marrow (BM) was harvested from the long bones of 6- to 8-week-old donor mice (FVB or C57BL/6N females), and hematopoietic stem and progenitor cells (HSPCs) were enriched by the depletion of lineage marker positive cells using a mix of biotinylated monoclonal antibodies against Gr1, Ter-119, CD45R, CD3 (1:15), and CD11b (1:2000) (BD Pharmingen) and the secondary reagents from the StemSep Mouse Progenitor Enrichment Kit (Stemcell Technologies). LV transduction was performed as previously described,18 and 106 cells were infused into the tail vein of lethally irradiated 6-week-old female C57BL/6N or FVB mice, or 5.5-week-old FVB/MMTV-PyMT mice. Radiation doses were 1100 cGy for FVB mice and 1300 cGy for C57BL/6N mice. The majority of experiments were done using the FVB strain, except those described in Figure 2C and supplemental Figures 2 and 3A, which used C57BL/6N mice.
Magnetic resonance imaging (MRI)
MRI examinations of PyMT mice were performed on a 3.0 Tesla human scanner (Achieva 3.0T; Philips), equipped with 80 mT/m gradients and a 40-mm volumetric coil (Micro-Mouse 40). The mice were anesthetized with Avertin and fixed in a prone position on a dedicated temperature control apparatus. A Turbo Spin Echo T2 (repetition time = 2500; echo time = 80; turbo factor = 9; field of view = 65 × 65 mm; zero-filled voxel size = 100 × 100 × 800 μm) sequence was acquired. Tumor volumes were calculated after manual segmentation of the lesion on the basis of signal intensity variation and enhancement characteristics by summing individual volumes (calculated as lesion area × slice thickness) of each slice.
Blood cell counts
Blood cell counts were determined using a Sysmex KX-21N apparatus. For each mouse, 10 μL of phosphate-buffered saline containing 45 mg/mL EDTA were added to 100 to 250 μL of peripheral blood.
Flow cytometry
For flow cytometry, we used a FACSCanto (BD Bioscience) platform. See supplemental Methods for further details.
Immunofluorescence staining and confocal microscopy
Tumors were cut into 12 to 16 µm cryostatic sections, as previously described.6 See supplemental Methods for further details.
Quantitative real-time polymerase chain reaction
Reactions were carried out in triplicate in an ABI Prism 7900HT Real-Time System (Applied Biosystems). For quantification of miRNA expression, small RNAs were extracted using the miRNeasy Mini Kit (Qiagen), and miRNA expression levels were determined by the Applied Biosystems TaqMan MicroRNA Assay system. miRNA expression was normalized to miR-16. Messenger RNA (mRNA) expression was quantified using either made-to-order TaqMan Gene Expression Assays (Applied Biosystems) or a custom-made TaqMan Low Density Array.9 See supplemental Methods for further details.
Statistical analysis
Unless otherwise noted, data are expressed as mean ± standard error of the mean (SEM). Statistical analyses were performed by Student t test, as indicated. Differences were considered statistically significant at P < .05.
Results
miR-155 expression in hematopoietic cells of normal and tumor-bearing mice
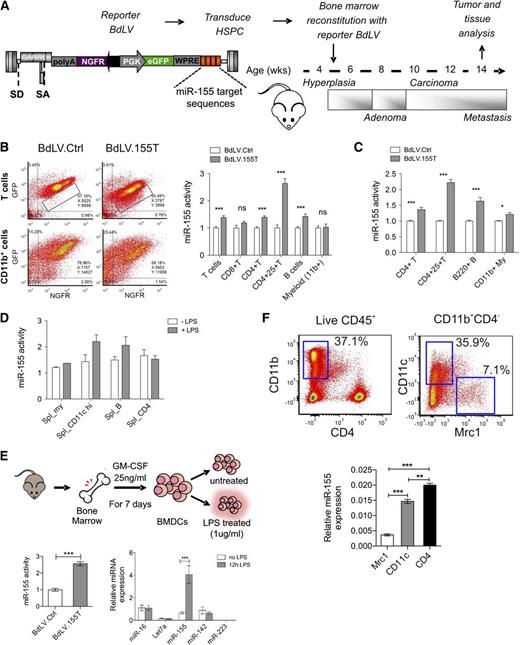
In order to investigate the expression and biological activity of miR-155 in the hematopoietic cells of tumor-bearing mice, we used a spontaneous breast cancer model (PyMT) closely mimicking tumor-host interactions.20 Before tumor onset, these mice were reconstituted with BM cells transduced with a bidirectional miR-155 reporter vector (BdLV.155T), which allows for the measurement of miR-155 activity in hematopoietic cells of various organs at single cell resolution16,21 (Figure 1A). In tumor-infiltrating hematopoietic cells, we detected significant miR-155 activity in B cells and CD4+ T cells, with the highest activity in regulatory T cells but not in CD8+ T cells or CD11b+ myeloid cells (Figure 1B). A similar picture was seen in the spleen (Figure 1C). However, we noted low but significant miR-155 activity also in CD11b+ myeloid cells, a cell subset that is progressively expanded with tumor growth. Because miR-155 is induced by inflammatory stimuli,22 we measured miR-155 activity in the splenocytes of BdLV.155T reconstituted wild-type mice that were or were not treated with lipopolysaccharide (LPS) (Figure 1D). LPS induced miR-155 activity, and this was most evident in CD11c+ APCs. A twofold to threefold induction of miR-155 activity could also be observed in vitro in primary BM-derived CD11c+ myeloid cells and in RAW264.7 cells after LPS stimulation, which corresponded to a sixfold induction of miR-155 expression as measured by quantitative polymerase chain reaction (qPCR) (Figure 1E; supplemental Figure 1). Thus, miR-155 activity is constitutive in CD4+ T cells and B cells and induced upon activation in CD11c+ APCs. Because the majority of TAMs infiltrating a late-stage tumor might not be in an activated stage, miR-155 activity might remain unnoticed when analyzing bulk CD11b+ cells. We thus sorted myeloid subpopulations from the CD45+ fraction of spontaneous mammary tumors taken from 13-week-old PyMT mice and measured the expression level of miR-155 by qPCR (Figure 1F).9 miR-155 was expressed in CD11c+ TAMs at similar levels to those in CD4+ T cells, but it was absent in the MRC1+ protumoral macrophages, suggesting that miR-155 is activated in a subset of TAMs with pro-inflammatory and antigen-presenting functions.
miR-155 expression in hematopoietic cell subsets of normal and tumor-bearing mice. (A) Experimental scheme: The BM of 5.5-week-old PyMT mice was reconstituted with wild-type FVB HSPCs transduced with miR-155 sensor (BdLV.155T) or Ctrl (BdLV.Ctrl) vectors. The chronology of breast cancer development and progression is indicated. (B) Representative dot plots show reporter expression (y-axis: GFP, miRNA reporter; x-axis: NGFR, normalizer) in tumor-infiltrating CD4+ lymphocytes (upper panel) or CD11b+ myeloid cells (lower panel) comparing BdLV.Ctrl (left) with BdLV.155T (right). Quantification of miR-155 activity in the indicated tumor-infiltrating hematopoietic subpopulations was performed as described in Materials and methods. Shown is the mean “fold repression” of BdLV.155T with respect to BdLV.Ctrl ± SEM, n = 7 to 12 mice. (C) miR-155 activity in splenocytes recovered from 13-week-old PyMT mice is shown, along with the mean fold repression of BdLV.155T with respect to BdLV.Ctrl ± SEM, n = 6 to 10 mice. (D) The graph shows miR-155 activity in splenocytes of wild-type mice with (gray) or without (white) in vivo LPS treatment (median ± range, n = 2). (E) Shown is the scheme of BM-derived dendritic cell (BMDC) differentiation protocol. miR-155 activity in BMDCs treated with LPS for 48 hours (redundant, since we stated in materials and methods the we show the mean +− sem unless otherwise noted n = 3) is shown in the graph (bottom left). qPCR of the indicated miRNAs in BMDCs that were or were not treated with LPS for 12 hours is shown in the graph (bottom right). The data show the relative abundance of each miRNA after normalization to miR-16 (n = 3). (F) Tumor-infiltrating hematopoietic cells were sorted from breast cancers of 13-week-old PyMT mice, as shown (top panels). miR-155 expression was measured in tumor-infiltrating macrophages (Mrc1+ or CD11c+ subsets) and CD4+ T lymphocytes by qPCR (bottom panel). Shown are the 2−ΔCt values using miR-16 as a normalizer (n = 6 from 2 independent experiments). **P < .01; *** P < .001. eGFP, enhanced green fluorescent protein; NGFR, truncated low affinity nerve growth factor receptor; PGK, phosphoglycerate kinase; SA, splice acceptor; SD, splice donor; WPRE, woodchuck hepatitis posttranscriptional regulatory element.
miR-155 expression in hematopoietic cell subsets of normal and tumor-bearing mice. (A) Experimental scheme: The BM of 5.5-week-old PyMT mice was reconstituted with wild-type FVB HSPCs transduced with miR-155 sensor (BdLV.155T) or Ctrl (BdLV.Ctrl) vectors. The chronology of breast cancer development and progression is indicated. (B) Representative dot plots show reporter expression (y-axis: GFP, miRNA reporter; x-axis: NGFR, normalizer) in tumor-infiltrating CD4+ lymphocytes (upper panel) or CD11b+ myeloid cells (lower panel) comparing BdLV.Ctrl (left) with BdLV.155T (right). Quantification of miR-155 activity in the indicated tumor-infiltrating hematopoietic subpopulations was performed as described in Materials and methods. Shown is the mean “fold repression” of BdLV.155T with respect to BdLV.Ctrl ± SEM, n = 7 to 12 mice. (C) miR-155 activity in splenocytes recovered from 13-week-old PyMT mice is shown, along with the mean fold repression of BdLV.155T with respect to BdLV.Ctrl ± SEM, n = 6 to 10 mice. (D) The graph shows miR-155 activity in splenocytes of wild-type mice with (gray) or without (white) in vivo LPS treatment (median ± range, n = 2). (E) Shown is the scheme of BM-derived dendritic cell (BMDC) differentiation protocol. miR-155 activity in BMDCs treated with LPS for 48 hours (redundant, since we stated in materials and methods the we show the mean +− sem unless otherwise noted n = 3) is shown in the graph (bottom left). qPCR of the indicated miRNAs in BMDCs that were or were not treated with LPS for 12 hours is shown in the graph (bottom right). The data show the relative abundance of each miRNA after normalization to miR-16 (n = 3). (F) Tumor-infiltrating hematopoietic cells were sorted from breast cancers of 13-week-old PyMT mice, as shown (top panels). miR-155 expression was measured in tumor-infiltrating macrophages (Mrc1+ or CD11c+ subsets) and CD4+ T lymphocytes by qPCR (bottom panel). Shown are the 2−ΔCt values using miR-16 as a normalizer (n = 6 from 2 independent experiments). **P < .01; *** P < .001. eGFP, enhanced green fluorescent protein; NGFR, truncated low affinity nerve growth factor receptor; PGK, phosphoglycerate kinase; SA, splice acceptor; SD, splice donor; WPRE, woodchuck hepatitis posttranscriptional regulatory element.
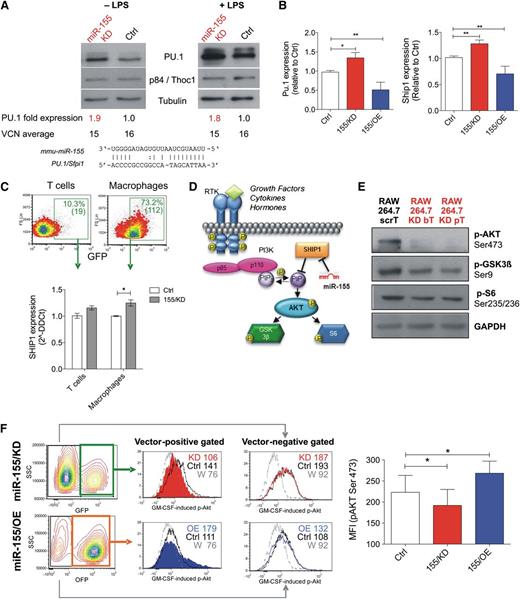
miR-155/KD upregulates natural miR-155 targets and reduces AKT activation. (A-B) Deregulation of natural miR-155 targets in RAW264.7 cells upon stable transduction with the indicated LVs is shown. (A) Western blot for PU.1 was performed on unstimulated and LPS-treated (1 μg/mL for 48 hours) cells. p84/Thoc1 and tubulin were used as loading controls; densitometric quantification of the PU.1 band after normalization to p84 and calibration to the scrambled control is shown below the blots, together with vector copy number in the tested cells. Blots are representative of 2 independent experiments. (B) qPCR for Pu.1 (left) and Ship1 mRNA (right) was performed. Data shown are the fold change to control, normalized to β2m (2−δδCt); n = 6 to 19 independent experiments. Statistical analysis of the data was performed on 2−ΔΔCt values by unpaired Student t test. (C) Shown are the results of qPCR for Ship1 mRNA in GFP+ T cells and macrophages sorted from the spleens of wild-type mice reconstituted with Ctrl (white) or 155/KD (gray) LV-transduced BMs 24 hours after treatment with 50 µg LPS intraperitoneally. n = 4 to 6 mice. (D) Shown is a simplified scheme of the PI3K/AKT signal transduction pathway. SHIP1, a direct miR-155 target, is a major negative regulator of the PI3K/AKT axis. (E) Phosphorylation of PI3K downstream targets in macrophage RAW264.7 cells transduced with ScrT Ctrl or miR-155/KD vectors (red); shown are western blots for pAKT (Ser473), pGSK3β (Ser9), and pS6 (Ser235/236). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as a loading control. (F) The panels show GM-CSF induced activation of AKT in RAW264.7 cells upon miR-155/KD (GFP+ cells) or miR-155/OE (OFP+ cells). The pAKT (S473) signal in vector-positive cells (middle histograms) and vector-negative cells (right histograms) is shown (FACS plots are representative). The colors represent the following: red, miR-155/KD vector; blue, 155/OE vector; black, Ctrl vector-transduced RAW264.7 cells; and gray, miR-155/KD cells pretreated with the PI3K inhibitor Wortmannin (W) to set the background signal. The MFI of each gated population is indicated. The column bar graph (right) shows pAKT Ser473 MFI in Raw264.7 cells transduced with the indicated vectors. Statistical analysis was performed on n = 4 (155/KD) or n = 3 (155/OE) replicates using a paired Student t test against the corresponding Ctrl vector-transduced samples. *P < .05; **P < .01. bT, bulged targets; MFI, mean flourescent intensity; pT, perfectly complementary targets; RTK, receptor tyrosine kinase; W, Wortmannin.
miR-155/KD upregulates natural miR-155 targets and reduces AKT activation. (A-B) Deregulation of natural miR-155 targets in RAW264.7 cells upon stable transduction with the indicated LVs is shown. (A) Western blot for PU.1 was performed on unstimulated and LPS-treated (1 μg/mL for 48 hours) cells. p84/Thoc1 and tubulin were used as loading controls; densitometric quantification of the PU.1 band after normalization to p84 and calibration to the scrambled control is shown below the blots, together with vector copy number in the tested cells. Blots are representative of 2 independent experiments. (B) qPCR for Pu.1 (left) and Ship1 mRNA (right) was performed. Data shown are the fold change to control, normalized to β2m (2−δδCt); n = 6 to 19 independent experiments. Statistical analysis of the data was performed on 2−ΔΔCt values by unpaired Student t test. (C) Shown are the results of qPCR for Ship1 mRNA in GFP+ T cells and macrophages sorted from the spleens of wild-type mice reconstituted with Ctrl (white) or 155/KD (gray) LV-transduced BMs 24 hours after treatment with 50 µg LPS intraperitoneally. n = 4 to 6 mice. (D) Shown is a simplified scheme of the PI3K/AKT signal transduction pathway. SHIP1, a direct miR-155 target, is a major negative regulator of the PI3K/AKT axis. (E) Phosphorylation of PI3K downstream targets in macrophage RAW264.7 cells transduced with ScrT Ctrl or miR-155/KD vectors (red); shown are western blots for pAKT (Ser473), pGSK3β (Ser9), and pS6 (Ser235/236). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as a loading control. (F) The panels show GM-CSF induced activation of AKT in RAW264.7 cells upon miR-155/KD (GFP+ cells) or miR-155/OE (OFP+ cells). The pAKT (S473) signal in vector-positive cells (middle histograms) and vector-negative cells (right histograms) is shown (FACS plots are representative). The colors represent the following: red, miR-155/KD vector; blue, 155/OE vector; black, Ctrl vector-transduced RAW264.7 cells; and gray, miR-155/KD cells pretreated with the PI3K inhibitor Wortmannin (W) to set the background signal. The MFI of each gated population is indicated. The column bar graph (right) shows pAKT Ser473 MFI in Raw264.7 cells transduced with the indicated vectors. Statistical analysis was performed on n = 4 (155/KD) or n = 3 (155/OE) replicates using a paired Student t test against the corresponding Ctrl vector-transduced samples. *P < .05; **P < .01. bT, bulged targets; MFI, mean flourescent intensity; pT, perfectly complementary targets; RTK, receptor tyrosine kinase; W, Wortmannin.
miR-155/KD upregulates endogenous miR-155 targets and attenuates AKT protein kinase signaling
In order to investigate whether miR-155 has a functional role in innate immune cells of tumor-bearing mice, we developed a lentiviral sponge vector (supplemental Figure 2A)18 that allows stable knockdown of miR-155 (miR-155/KD) in myeloid cells, but it has little activity in T lymphocytes. To this aim, we exploited the spleen focus forming virus (SFFV) promoter, which drives transgene expression to high levels in monocytes and dendritic cells, to intermediate levels in granulocytes and B lymphocytes, and to very low levels in T lymphocytes, as shown in vivo after BM reconstitution by HSPCs transduced with green fluorescent protein (GFP) expressing SFFV LVs containing scrambled (Ctrl) or miR-155 sponge sequences (155/KD) (supplemental Figure 2B-C). The miR-155/KD vector was tested for its ability to knock down miR-155 in RAW264.7 cells, which show basal miR-155 expression levels similar to inflammatory TAMs (see Figure 1F; supplemental Figure 1A). We measured the expression levels of PU.1 protein, a previously reported miR-155 target,23 in RAW264.7 cells after transduction with multiple copies of the miR-155/KD or Ctrl LV (Figure 2A). Both in steady state conditions and upon LPS treatment, PU.1 was upregulated up to twofold times in miR-155/KD cells as measured by western blot analysis (Figure 2A). Similarly, reverse transcription qPCR for Pu.1 and Ship1 mRNA, another well-established miR-155 target,24 showed an ∼1.5-fold upregulation upon miR-155/KD, while overexpression of miR-155 (miR-155/OE) using a previously described LV design19 resulted in a 1.5 to twofold downregulation (Figure 2B). Importantly, we showed a significant upregulation of Ship1 mRNA in splenic macrophages of mice reconstituted with SFFV.155T/KD LV–transduced cells as opposed to SFFV.Ctrl LV–transduced cells after in vivo LPS challenge (Figure 2C). This result confirms the inhibition of miR-155 regulation in myeloid cells in vivo by our vector. Whereas the majority of splenic macrophages expressed high levels of GFP, indicating strong expression of the sponge transcript and functional inhibition of miR-155, only a minority of T cells expressed GFP to barely detectable levels, consistently with low expression of the sponge promoter and functional activity of miR-155 in these cells. We then sorted and analyzed the subset of GFP-expressing T cells and detected a minor modulation of the Ship1 target, which did not reach statistical significance. These data confirmed that, even in the minority of T cells in which sponge expression may have been enhanced by integration site-specific effects or high vector copy number, we did not achieve robust miR-155 knockdown. Thus, we conclude that our vector allows miR-155/KD in vivo specifically in myeloid cells. We then investigated the impact of miR-155/KD on signaling pathways associated with Ship1, a major negative regulator of growth factor receptor-mediated PI3K/AKT activation and myeloid cell survival (Figure 2D). Western blot analysis of RAW264.7 cells indicated reduced levels of AKT phosphorylation at Ser473, a major activatory phosphorylation site, upon miR-155/KD (Figure 2E). This was accompanied by a reduction in phosphorylation of GSK3β at Ser9 and of S6 kinase at Ser235/236, 2 substrates of AKT. Thus, miR-155/KD impairs AKT activation in macrophages by increasing Ship1 levels. Next we tested whether the response of macrophages to granulocyte monocyte colony stimulating factor (GM-CSF), a pro-inflammatory cytokine involved in the activation and maturation of APC signaling through the PI3K/AKT pathway, was altered upon miR-155/KD or OE. GM-CSF–induced AKT (Ser473) phosphorylation as assessed by phosphoflow was reduced in miR-155/KD cells and enhanced in miR-155/OE cells, and these effects were only evident when gating on the transduced cell population (Figure 2F). These data indicate that miR-155/KD alters signaling pathways known to have a crucial function in inflammatory macrophage activation.
Myeloid-specific miR-155 knockdown accelerates tumor growth in a spontaneous breast cancer model
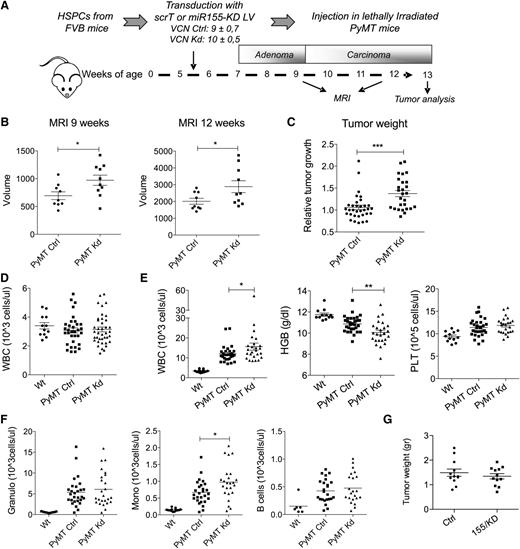
Next we addressed the question of whether miR-155 has a functional role in TAMs. To this aim, we transplanted PyMT mice with HSPC transduced with multiple copies of the miR-155/KD vector (155/KD mice) or the Ctrl LV (Ctrl mice) (Figure 3A). At the time of transplant (5.5 weeks of age), PyMT mice showed hyperplasia of the breast epithelium with no detectable adenomas or carcinomas.25 Spontaneous tumor development was monitored by MRI at 9 and 12 weeks of age (Figure 3B). Strikingly, miR-155/KD in BM-derived cells significantly increased tumor volume compared with the control group, and this effect was already evident at the late adenoma stage (9-week time point). The early appearance of the effect further supports a myeloid cell–mediated phenotype, because T-cell reconstitution had not yet occurred 3.5 weeks after HSPC transplantation, in sharp contrast to myeloid lineage cells that show near complete donor chimerism at 2 weeks (supplemental Figure 3A). In fact, ∼50% of myeloid cells were miR-155/KD (estimated by GFP expression) at the time of the first MRI, while <0.7% of T cells expressed low levels of GFP (supplemental Figure 3B). Mice were euthanized between 12 and 13 weeks of age, and accelerated tumor growth in the miR-155/KD group was confirmed by the significantly increased weight of the surgically resected tumor masses (Figure 3C). Next we assessed the impact of miR-155/KD on hematopoiesis in PyMT mice. Blood cell counts did not show significant differences early after transplant, suggesting that miR-155/KD had no evident impact on hematopoietic reconstitution (Figure 3D). In addition, wild-type mice reconstituted with BM cells transduced with the SFFV.155T/KD vector and observed for 5 months showed no gross alterations in hematopoietic lineage counts in the peripheral blood, BM, spleen, and thymus, with the exception of a slight increase in B cells in the BM and spleen but not in the peripheral blood (supplemental Figure 2D-F). A similar phenotype was observed in miR-155−/− mice,13 further confirming efficient miR-155 knockdown by our LV construct. Taken together, these data suggest that loss of miR-155 has no evident impact on hematopoietic reconstitution or steady state myelo-, erythro-, and megakaryopoiesis. However, in 12-week-old PyMT mice, tumor-induced anemia and leukocytosis were significantly more severe in the miR-155/KD group compared with the scrambled Ctrl group (Figure 3E). Among the leukocytes, side scatter (SSC)low CD11b+ monocytes showed the most significant increase in miR-155/KD vs Ctrl mice (Figure 3F). These monocytes have been reported to be protumoral and proangiogenic.26,27 To investigate whether miR-155/KD in myeloid cells would also accelerate the growth of established carcinomas, we set up an orthotopic breast cancer model. Tumors from 14-week-old PyMT mice (late carcinoma stage) were dissociated and orthotopically injected into the mammary fat pad of FVB mice stably reconstituted with HSPCs expressing miR-155/KD or Ctrl vector. Even though hematopoietic reconstitution with miR-155/KD cells was as high as that obtained in the PyMT mice, we observed no significant differences in tumor growth between the miR-155/KD and Ctrl groups (Figure 3G). In summary, miR-155/KD accelerates tumor development in a spontaneous mammary cancer model acting during the early stages of carcinogenesis by a mechanism involving myeloid cells.
miR-155 knockdown in myeloid cells accelerates tumor growth. (A) Experimental scheme: The BM of 5.5-week-old PyMT mice was reconstituted with wild-type FVB HSPCs that were transduced with the miR-155/KD or ScrT Ctrl vectors. Mean vector copy numbers per cell ± SEM are indicated. Chimeras are referred to as PyMT Kd mice and PyMT Ctrl mice. (B) Mammary tumor growth was measured by MRI (mean volume ± SEM of the 3 anterior mammary gland pairs) at 9 and 12 weeks of age in PyMT Ctrl (n = 9) and PyMT Kd (n = 10) mice. (C) Tumor burden at the experimental end point (13 weeks) was determined by weighing the mass of surgically resected tumor specimens from each mouse. Shown is the mean relative tumor weight in PyMT Ctrl mice (n = 33) and PyMT Kd mice (n = 27). Within each of 4 independent experiments, tumor mass was normalized to the median tumor weight of the respective PyMT Ctrl group. (D) White blood cell (WBC) count was measured in wild-type FVB mice (n = 12), PyMT Ctrl (n = 33), and PyMT Kd mice (n = 39) at 9 weeks of age. (E) Complete blood cell counts and hemoglobin levels were measured in wild-type FVB mice (n = 12), PyMT Ctrl (n = 30), and PyMT Kd mice (n = 25) at 13 weeks of age. Results are from 4 independent experiments. (F) Differential WBC counts were obtained by multiplying the absolute WBC shown in (E) by the fraction of CD11b+side scatter (SSC)hi cells (granulocytes, left panel), CD11b+SSClow cells (monocytes, middle panel), and CD19+ cells (B lymphocytes, right panel) as measured by flow cytometry. (G) The graph shows tumor growth in FVB mice transplanted with miR-155/KD or Ctrl vector–transduced HSPCs and orthotopically challenged with late-stage PyMT mammary tumor cells 6 weeks after transplantation. Shown is the mean tumor weight ± SEM; n = 6 mice per group. HGB, hemoglobin; PLT, platelets; Wt, wild type.
miR-155 knockdown in myeloid cells accelerates tumor growth. (A) Experimental scheme: The BM of 5.5-week-old PyMT mice was reconstituted with wild-type FVB HSPCs that were transduced with the miR-155/KD or ScrT Ctrl vectors. Mean vector copy numbers per cell ± SEM are indicated. Chimeras are referred to as PyMT Kd mice and PyMT Ctrl mice. (B) Mammary tumor growth was measured by MRI (mean volume ± SEM of the 3 anterior mammary gland pairs) at 9 and 12 weeks of age in PyMT Ctrl (n = 9) and PyMT Kd (n = 10) mice. (C) Tumor burden at the experimental end point (13 weeks) was determined by weighing the mass of surgically resected tumor specimens from each mouse. Shown is the mean relative tumor weight in PyMT Ctrl mice (n = 33) and PyMT Kd mice (n = 27). Within each of 4 independent experiments, tumor mass was normalized to the median tumor weight of the respective PyMT Ctrl group. (D) White blood cell (WBC) count was measured in wild-type FVB mice (n = 12), PyMT Ctrl (n = 33), and PyMT Kd mice (n = 39) at 9 weeks of age. (E) Complete blood cell counts and hemoglobin levels were measured in wild-type FVB mice (n = 12), PyMT Ctrl (n = 30), and PyMT Kd mice (n = 25) at 13 weeks of age. Results are from 4 independent experiments. (F) Differential WBC counts were obtained by multiplying the absolute WBC shown in (E) by the fraction of CD11b+side scatter (SSC)hi cells (granulocytes, left panel), CD11b+SSClow cells (monocytes, middle panel), and CD19+ cells (B lymphocytes, right panel) as measured by flow cytometry. (G) The graph shows tumor growth in FVB mice transplanted with miR-155/KD or Ctrl vector–transduced HSPCs and orthotopically challenged with late-stage PyMT mammary tumor cells 6 weeks after transplantation. Shown is the mean tumor weight ± SEM; n = 6 mice per group. HGB, hemoglobin; PLT, platelets; Wt, wild type.
miR-155 knockdown impairs the ability of myeloid cells to mount a pro-inflammatory response
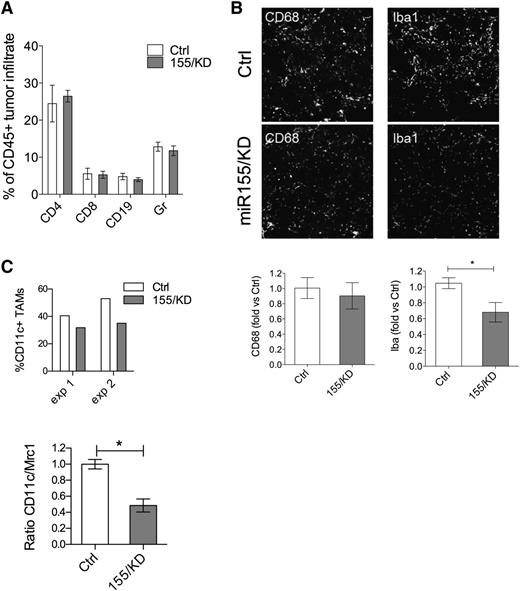
Tumor-infiltrating hematopoietic cells are important effectors of tumor rejection processes, especially during early-stage tumorigenesis, before tumor cells have acquired mechanisms of immune evasion.28 We quantified the composition of the inflammatory cell infiltrate in 12-week-old PyMT tumors from either miR-155/KD or Ctrl mice. The frequency of infiltrating CD45+ hematopoietic cells was similar in tumor sections of miR-155/KD and control mice (data not shown), and flow cytometric analysis of dissociated tumors showed no differences in the relative proportion of Gr1+ cells, CD4+, or CD8+ T and CD19+ B lymphocytes (Figure 4A). Likewise, CD68+ total macrophages were equally represented in the 2 groups (Figure 4B). However, there was a significant reduction in the expression of the activation marker ionized binding adaptor molecule-1 (IBA-1) by innate immune cells in miR-155/KD tumors (Figure 4B), and the myeloid infiltrate comprised fewer CD11c+ cells as assayed by flow cytometry and qPCR (Figure 4C). These data suggest that miR-155/KD impairs activation of inflammatory macrophages in the tumor. We then performed gene expression analysis on FACS-sorted, GFP+CD11b+F4/80+ TAMs isolated from PyMT mammary tumors by using a custom-made TaqMan Low Density Array carrying 95 genes including cytokines, chemokines, and genes involved in inflammation and immune response (supplemental Table 1). Three genes were differentially expressed between the miR-155/KD and Ctrl groups: interleukin (Il)-12b was significantly downregulated, while Il-4 and Il-13 were significantly upregulated in the miR-155/KD myeloid infiltrate (Table 1). The same low density array was run on CD4+ tumor-infiltrating lymphocytes (supplemental Table 2). Interestingly, Gm-csf and Il-13 were both significantly upregulated in T cells from the miR-155/KD group, whereas Il-6 and Ccr6 were significantly downregulated (Table 2). These data support a skewing of the tumor immune microenvironment from a classic pro-inflammatory activation state to an alternatively activated phenotype comprising a Th2 profile. Collectively, these data highlight an impaired capacity of miR-155–deficient innate immune cells in establishing a pro-inflammatory microenvironment within the tumor, likely resulting in a reduction of antitumor immune responses.
miR-155 knockdown in myeloid cells reduces TAMs with pro-inflammatory activity. (A) The composition of hematopoietic infiltrate in PyMT breast cancer was determined by flow cytometric analysis of CD45+ cells derived from dissociated tumors. The proportion of Gr1+ cells, CD4+ or CD8+ T lymphocytes, and CD19+ B lymphocytes within the total CD45+ cells is shown (mean ± SEM, n = 9-10). (B) Shown is representative immunofluorescence microscopy of tumor sections (top panels) from 13-week-old PyMT Kd or PyMT Ctrl mice stained for the pan-macrophage marker CD68 and IBA-1, a marker associated with macrophage activation. Original magnification ×20. Shown is the quantification of the CD68+ (bottom, left panel) and IBA-1+ (bottom, right panel) areas from PyMT tumor sections relative to PyMT Ctrl (n = 6 mice from 2 independent experiments). (C) The graph (upper panel) depicts the fraction of CD11c+ TAMs within the total CD45+ hematopoietic tumor infiltrate as measured by flow cytometry. Shown are 2 independent experiments; each point is a pool of 6 and 3 mice in experiment 1 and experiment 2, respectively. The graph lower panel shows the ratio of CD11c/Mrc1 transcript in the CD11b+F4/80+ TAM population as measured by qPCR (n = 3 pools of 3 mice each). Statistical analyses were performed by Student t test. * P < .05.
miR-155 knockdown in myeloid cells reduces TAMs with pro-inflammatory activity. (A) The composition of hematopoietic infiltrate in PyMT breast cancer was determined by flow cytometric analysis of CD45+ cells derived from dissociated tumors. The proportion of Gr1+ cells, CD4+ or CD8+ T lymphocytes, and CD19+ B lymphocytes within the total CD45+ cells is shown (mean ± SEM, n = 9-10). (B) Shown is representative immunofluorescence microscopy of tumor sections (top panels) from 13-week-old PyMT Kd or PyMT Ctrl mice stained for the pan-macrophage marker CD68 and IBA-1, a marker associated with macrophage activation. Original magnification ×20. Shown is the quantification of the CD68+ (bottom, left panel) and IBA-1+ (bottom, right panel) areas from PyMT tumor sections relative to PyMT Ctrl (n = 6 mice from 2 independent experiments). (C) The graph (upper panel) depicts the fraction of CD11c+ TAMs within the total CD45+ hematopoietic tumor infiltrate as measured by flow cytometry. Shown are 2 independent experiments; each point is a pool of 6 and 3 mice in experiment 1 and experiment 2, respectively. The graph lower panel shows the ratio of CD11c/Mrc1 transcript in the CD11b+F4/80+ TAM population as measured by qPCR (n = 3 pools of 3 mice each). Statistical analyses were performed by Student t test. * P < .05.
Discussion
Here we describe a key role of miR-155 in mediating the antitumor responses of myeloid cells in vivo. By knocking down miR-155 in the myeloid compartment of transgenic mice that spontaneously develop breast cancer, we observed (1) an acceleration of tumor growth accompanied by a more severe tumor-induced anemia and monocytosis; (2) a significant reduction in the proportion of CD11c+ inflammatory TAMs and in the expression of the macrophage activation marker IBA-1; and (3) a skewing of the cytokine milieu of the tumor toward an M2/Th2 phenotype evident in the gene expression profile of both TAMs and CD4+ TILs (tumor-infiltrating lymphocytes). Contrary to the prevailing view that myeloid cells favor tumor growth and progression, our finding highlights an important role of hematopoietic cells in constraining tumor development and growth. By antagonizing an miRNA exclusively in hematopoietic cells, a comparably minor manipulation, we obtained a conspicuous acceleration of tumor growth in an already highly aggressive spontaneous breast cancer model.
Several lines of evidence suggest that the observed acceleration of tumor growth is mediated by miR-155 insufficiency in myeloid cells and, particularly, in APCs such as inflammatory macrophages/dendritic cells. These include the myeloid-biased expression of the knockdown vector, the occurrence of tumor acceleration before the onset of T-cell reconstitution following HSPC transplantation, and the changes in TAM phenotypes such as fewer CD11c+ cells and the reduced expression of the activation marker IBA-1. On the contrary, we expect little/no influence of miR-155/KD on the protumoral macrophage subsets because they do not express relevant levels of miR-155.
We investigated the molecular consequences of miR-155/KD in a macrophage cell line and in primary hematopoietic cells in vivo after BM reconstitution. We found an upregulation of Ship1 and, consequently, a reduction in the activation of the PI3K/AKT pathway at baseline and upon exposure to pro-inflammatory stimuli such as LPS and GM-CSF. SHIP1 is an inositol phosphatase and, together with PTEN, the main negative regulator of PI3K/AKT signaling.29,30 It represents 1 of the main direct targets of miR-15531 and represses the production of pro-inflammatory cytokines, phagosome maturation, and macrophage survival.32 Consistently, AKT promotes the production of pro-inflammatory cytokines such as TNF-α, IL-6, and IL-12, while inhibiting the production of the anti-inflammatory cytokine IL-10.32 This might provide a mechanistic explanation for the reduced occurrence of classically activated myeloid cells in miR-155/KD tumors and the M2/Th2-biased cytokine profile that we observed in the tumor. In CD4+ TILs, we found a significant upregulation of Il-13 and Gm-csf, and we found a downregulation of Il-6, Csf1r, and Pu.1 in the miR-155/KD group. IL-13 is a classic Th2 cytokine. In addition, downregulation of PU.1 might further reinforce Th2 functions because Pu.1−/− mice have been shown to have a lower T-cell receptor activation threshold under Th2 culture conditions.33 Instead, IL-6, which we found significantly downregulated in the miR-155/KD group, is a classic pro-inflammatory cytokine.34 Macrophages and T cells interact closely, and it has been suggested that they mutually polarize each other to a pro-inflammatory M1/Th1 phenotype on one end of the spectrum, or to an alternative M2/Th2 activation state on the other end.35 Thus, the Th2 bias within the T-cell compartment might indirectly reflect a functional impairment of pro-inflammatory macrophages to induce Th1 polarization, causing an imbalance toward pro-angiogenic, protumoral TAMs and a Th2 phenotype. Our gene expression data, which show increased Il-4 and Il-13 and decreased Il-12b levels in TAMs upon miR-155/KD, further support a shift from M1 to M2 polarized macrophages. Recent reports suggested that miR-155 can directly modulate macrophage phenotypes. Arranz et al36 showed that a reduction in miR-155 directly induces an M2 macrophage profile in mouse peritoneal macrophages by upregulating C/EBPβ, a direct miR-155 target and a key regulator of arginase 1 expression. Moreover, ectopic miR-155 expression can repolarize protumoral M2 TAMs toward an antitumoral M1 phenotype,37 and augmenting miR-155 levels in tumor-associated dendritic cells by miRNA mimetics enhanced antitumor responses in established ovarian cancers.38 These observations are fully in line with our data, which show impairment of classic macrophage activation upon miR-155/KD. We do not exclude, however, that miR-155 insufficiency in other hematopoietic compartments may have similar protumoral effects, as recently proposed for T cells by Huffaker et al.39
It has been suggested that miR-155 positively regulates granulocyte and monocyte development by acting on myeloid BM progenitors,12 a compartment in which the SFFV-driven miR-155/KD vector is active.18 However, our in vivo results show an increase in the peripheral myeloid compartment (CD11b+ monocytes) upon miR-155/KD. Thus, it is more likely that the expanded myeloid compartment is a consequence of the increased tumor burden upon myeloid-specific miR-155/KD rather than a direct effect of the miR-155/KD vector on the BM compartment. Indeed, wild-type mice showed no alterations in blood cell counts when transplanted with miR-155/KD LV–transduced hematopoietic progenitors. In support of tumor-driven myeloid expansion, we noted increased GM-CSF expression in CD4+ TILs from the miR-155/KD group as compared with the control. GM-CSF is 1 of the principal myeloid growth factors stimulating myelopoiesis and recruiting myeloid cells to the tumor.40 An additional mechanism of monocyte mobilization might involve angiotensin receptor 1, which has been reported as a direct miR-155 target and is supposed to be upregulated upon miR-155/KD.41,42
Extensive data indicate that miR-155 functions as an oncomiR in several solid as well as hematologic tumors,43,44 and it is generally considered a marker of poor prognosis.45,46 Constitutive miR-155 expression in hematopoietic cells can result in the development of hematologic malignancies such as lymphoma and myeloproliferative disease.12,45,47 These evidences have motivated the development of anti–miR-155 drugs as potential anticancer therapeutics.43,48-50 Our data indicate that the oncogenic activity of miR-155 is cell context dependent, and thus suggest some caution before applying anti–miR-155 treatments to cancer patients. In this regard, it should be kept in mind that miR-155 has a physiological role in orchestrating a productive immune response. Previous work has highlighted the importance of miR-155 in acute inflammatory insults.13,14,22 Our data expand the role of miR-155 to myeloid cell–mediated antitumoral responses, because blocking its physiological upregulation in these cells accelerated tumor development in a spontaneous breast cancer model. Thus, a wave of miR-155 activation in differentiated hematopoietic cells boosts immunity, and only failure to downregulate miR-155 may support oncogenesis, suggesting that miR-155 is, first of all, an “immunomiR” rather than an oncomiR. It remains to be determined whether the potential benefits of miR-155 inhibition (ie, reduction in cell autonomous tumor cell growth in selected cancers with miR-155 overexpression) outweigh immunosuppression, which can, paradoxically, favor tumor growth.
In summary, we here describe an antitumoral role of miR-155 as an integral effector of immunosurveillance, thus limiting early steps of breast cancer development. This work highlights the context specificity and complexity of miRNA regulation, calling for a cell type specific approach when targeting miRNA function, which takes into consideration the heterogeneity of tumors composed of both cancer cells and a collection of nonmalignant host cells. Moreover, our findings support the concept that TAMs make up distinguishable subsets within the tumor, which mediate different and sometimes opposing functions in tumor development and growth.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Alice Giustacchini, Maura Rossetti, Andrea Annoni, and Nada Sonda for help with some experiments; Alessio Palini for cell sorting; Clelia Di Serio for the original design of the statistical analysis for the low density array; Lucia Sergi Sergi for vector production; and Giulia Schira, Anna Ranghetti, and Davide Moi for technical help with some experiments. The authors also thank Michele De Palma and Vincenzo Bronte for helpful discussions.
This work was supported by grants from the Italian Association for Cancer Research and the European Union (ERC Advanced Grant 249845, TARGETINGGENETHERAPY) (L.N.). E.Z. was supported by a fellowship from the Italian Foundation for Cancer Research.
Authorship
Contribution: E.Z. designed and performed research, analyzed data, and drafted the manuscript; F.P. analyzed gene expression data; M.S. performed western blot and phosphoflow analyses; R.M. discussed the data; L.S.P. performed MRI; and L.N. and B.G. conceived and coordinated the research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Luigi Naldini, San Raffaele Telethon Institute for Gene Therapy, Via Olgettina, 58, 20132 Milan, Italy; e-mail: naldini.luigi@hsr.it.
References
Author notes
B.G. and L.N. share senior authorship.