Abstract
Although T-cell cytokines are believed to play an essential role in the development of acquired aplastic anemia (AA), it remains unclear which cytokines or cytokine signals trigger hematopoietic stem cell (HSC) suppression in AA. Gene expression profiling of bone marrow (BM) T cells may be useful for identifying inciting molecules, but the BM T cells in patients with severe AA are inappropriate subjects for such studies, because their gene expression profile suffers changes due to the T-cell exposure to various cytokines. Some patients with moderate AA present with profound thrombocytopenia and an increase in glycosylphophatidylinositol-anchored protein (GPI-AP)-deficient cells without significant neutropenia and anemia, and eventually progress to pancytopenia that fulfills the criteria of AA. They usually show a rapid and complete response to cyclosporine (CsA) if treated early after the thrombocytopenia is manifested. Such patients with the “pre-AA” state may serve as ideal subjects to clarify which genes are involved in the development of AA.
To identify the genes closely associated with the development of AA responsive to CsA, we carried out gene expression profiling of the BM mononuclear cells (BMMCs) obtained from 6 un-transfused patients with BM failure with increased GPI-AP-deficient cells (two with the pre-AA state and four with moderate AA) and three healthy individuals using a microarray analysis with Agilent Microarrays, and identified genes that were highly expressed in patients' BMMCs at the diagnosis. All patients responded to CsA monotherapy and achieved platelet recovery≥50 x 109/L within one year.
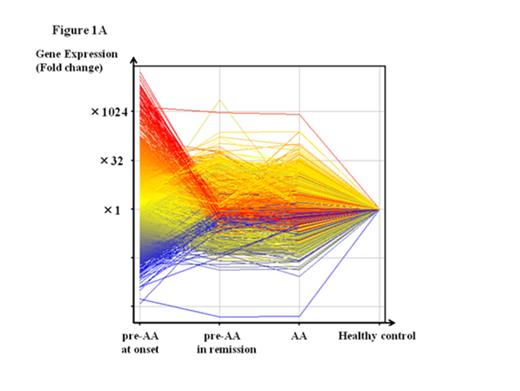
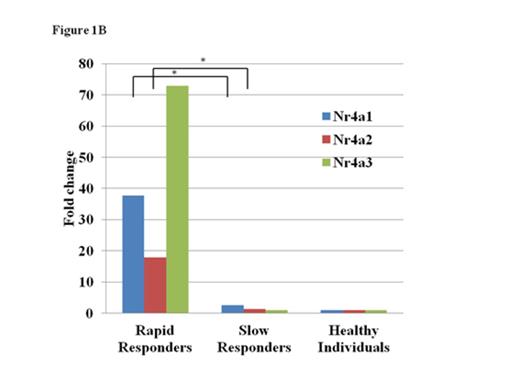
A total of 1119 genes, including IL17C, CD40LG and NR4A1, were upregulated in the BMMCs of the six patients at onset by at least two-fold the level of healthy individuals. The extent of gene overexpression was more pronounced in two patients with the pre-AA state than in those with MAA (Fig. 1A). A comparison of the gene expression levels in BMMCs at onset with those in BMMCs obtained after achieving the platelet count≥100 x 109/L in two rapid responders to CsA revealed 1858 genes, including TNFSF9, NFATC2 and CCL4, that were upregulated. Of note, all three Nr4A family members, including Nr4a1 (Nur77), Nr4a2 (Nurr1) and Nr4a3 (NOR-1), were upregulated at onset (17.4-, 46.5- and 106.4-fold increases compared to those after CsA therapy; 37.7-, 17.9- and 72.9-fold increases compared to healthy controls, respectively) in patients who obtained the good platelet response within three months of CsA therapy (rapid responders). The expression levels of Nr4A genes in four AA patients who showed a slow response to CsA (slow responders) were comparable to those of healthy individuals (Fig. 1B).
Genes in the Nr4a subfamily are immediate early genes that can be induced by a wide array of stimuli, including growth factors and inflammatory signals. Nr4a2 is reportedly upregulated in T cells of patients with multiple sclerosis via the calcineurin–NFAT pathway, and plays a pivotal role in mediating cytokine production, such as the production of IL-17 from pathogenic T cells. In patients with early stage AA, some inflammatory stimuli may induce the overexpression of Nr4A, leading to the development of BM failure. Quantification of the Nr4A expression of BMMCs may therefore be useful for diagnosing thrombocytopenia that can progress to AA and also predicting a rapid response to CsA therapy in patients with the early stage AA.
Nakao:Alexion: Honoraria, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.