Abstract
The exact disease state of chronic myeloid leukemia (CML) patients in deep molecular remission is unknown, since even the most sensitive quantitative polymerase chain reaction (qPCR) methods cannot identify the patients who are prone to relapse after treatment withdrawal. Consequently, new strategies are needed to unravel the true disease state in these patients. Through evaluation of FACS sorted stem- and progenitor cells with standard qPCR and interphase fluorescence in situ hybridization (iFISH) assays, we here demonstrate that residual disease can be assessed at the stem cell level in patients with 4-log reduction in BCR-ABL expression (MR4).
Fresh bone marrow aspirates were acquired from 17 CML patients treated with tyrosine kinase inhibitors (TKIs) for a period of 12-119 months. Samples were enriched for CD34+ cells, stained with fluorescent monoclonal antibodies and FACS sorted. In the sort setup we included the human myeloid inhibitory C-type lectin-like receptor (hMICL) antigen (van Rhenen et al Blood 2007, Larsen et al Cytometry 2012), which identifies granulocyte-macrophage committed progenitors. Sorted subsets were: Hematopoietic stem cells (HSCs, CD34+CD38-), hMICL+ progenitors (MpP; CD34+CD38+hMICL+), and hMICL- progenitors (MnP; CD34+CD38+hMICL-). Sorted cells were evaluated for their content of Philadelphia positive (Ph+) cells using both iFISH and a sensitive qPCR assay optimized to small cell samples.
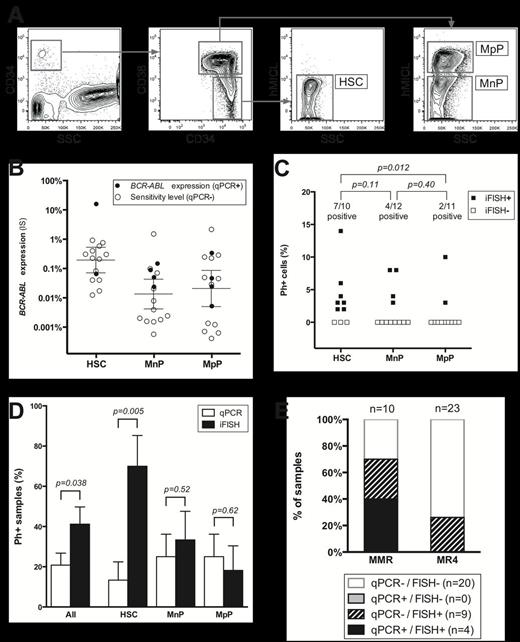
Detection of residual Ph+ cells in FACS sorted CD34+ cell subpopulations in CML patients in MMR and MR4. (A) FACS sorting of HSCs, MpPs, and MnPs. (B) Expression of BCR-ABL mRNA (on the International Scale (IS)) in the sorted CD34+ subsets. Error bars denote the median upper boundary of BCR-ABL expression (± 95% confidence interval). (C) Residual Ph+ cells as detected by iFISH in the sorted CD34+ subsets. (D) Detection of residual Ph+ cells in sorted subsets by iFISH and mRNA-based qPCR. Error bars denote SEM. (E) Combined qPCR and iFISH status of samples analyzed by both modalities.
Detection of residual Ph+ cells in FACS sorted CD34+ cell subpopulations in CML patients in MMR and MR4. (A) FACS sorting of HSCs, MpPs, and MnPs. (B) Expression of BCR-ABL mRNA (on the International Scale (IS)) in the sorted CD34+ subsets. Error bars denote the median upper boundary of BCR-ABL expression (± 95% confidence interval). (C) Residual Ph+ cells as detected by iFISH in the sorted CD34+ subsets. (D) Detection of residual Ph+ cells in sorted subsets by iFISH and mRNA-based qPCR. Error bars denote SEM. (E) Combined qPCR and iFISH status of samples analyzed by both modalities.
Detection of residual disease in the CD34- and in at least one of the analyzed CD34+ fractions of the individual patients as assessed by both iFISH and mRNA-based qPCR.
Detection of residual disease in the CD34- and in at least one of the analyzed CD34+ fractions of the individual patients as assessed by both iFISH and mRNA-based qPCR.
To our knowledge, we are the first to directly demonstrate the discrepancy between DNA- and mRNA-based methods when assessing persisting Ph+ stem- and progenitor cells in patients with MR4. Pre-selection of CD34+ stem- and progenitor cells unmasked residual disease, and enabled detection of Ph+ cells by iFISH in almost all analyzed patients, including 5/6 of patients with MR4. Our results show that the residual Ph+ cells in CML patients with deep molecular response are low BCR-ABL producers, and that DNA-based methods are required to define the next level of response beyond the achievement of undetectable BCR-ABL mRNA. This approach demonstrates that residual disease can be assessed at the stem cell level in CML patients with MR4, and may provide us with a tool for early and safe identification of candidates for TKI withdrawal.
Hokland:Novartis: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.