Abstract
T cell repertoire in health and disease is complex, comprised of millions of unique T cell clones defined by hetero-dimeric T cell receptors (TCR). The a and b subunits of the TCR are products of genetic loci consisting of variable (V) and joining (J) segments in the case of TCR a (TRA), and V, J and diversity (D) segments in the case of TCR b (TRB). These gene segments are recombined to yield TCR a (VJ) or TCR b (VDJ). Recently we reported that T cell clonal frequency in normal stem cell donors, from the point of view of TCR b gene segment usage rather than being random, has a fractal ordering (Meier et al, BBMT 2013). Fractals are mathematical constructs characterized by self-similarity at different scales of measurement. Thus whether T cell repertoire is examined from the standpoint of DJ, VDJ or VDJ+nucleotide inserted TCR b clones the frequency distribution remains the same when log transformed data are examined yielding a fractal dimension (FD) of ~ 1.5.
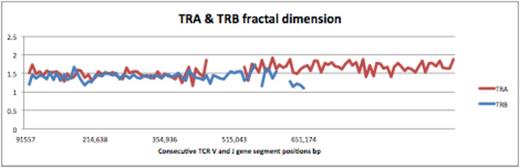
TCR a (TRA) and b (TRB) fractal dimension calculated for each variable and joining segment (TCR d region on the TRA locus is excluded)
TCR a (TRA) and b (TRB) fractal dimension calculated for each variable and joining segment (TCR d region on the TRA locus is excluded)
During T cell receptor recombination, the J and the D segments in TRA and TRB loci respectively, are recombined with the V segments therefore the distribution of each V segment in the TRA and TRB loci was determined relative to the position of the J and the D segments in the respective loci, by the using the following formula spacing V-J or V-D = INP-D or J segment / INP nthV segment, (INP: initial nucleotide positions of gene segments). This measure demonstrated a strikingly consistent proportionality across both TRA and TRB loci, and when plotted for successive V segments on the TRB locus, the value declined logarithmically, with a slope of approximately 1.6, and similarly an average slope of 1.3 for the TRA locus, consistent with the earlier observed FD for these loci. We conclude that T cell receptor gene segments are organized in an iterating logarithmically scaled manner. This implies that aside from mechanisms such as the 12/23 rule, gene segment distribution across the T cell receptor loci may help determine the order of recombination, and thus contribute to the observed fractal ordering of the T cell repertoire.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.