Abstract
Immunoglobulin light chain-associated amyloidosis (AL) is caused by deposition of immunoglobulin light chain molecules with unique, clonotypic variable (V) regions. Detection and identification of the V region, however, has been challenging due heterogeneity inherent in V regions. We developed a new method for detecting IGKV and IGLV region fragments in AL deposits using protein mass spectrometry and novel bioinformatics approach. We report distinct IGKV and IGLV usage in localized versus systemic AL.
Shotgun protein mass spectrometry on amyloid plaques was performed as previously described (Blood. 2009; 114: 4957-9). Peptide tandem mass spectra (MS/MS) were searched against a composite sequence database containing SwissProt's human complete proteome augmented with 1764 IG V region sequences obtained from ImMunoGeneTics database and a Mayo Clinic internal database. Reversed sequences were appended to the database for estimating identification false discovery rates (FDRs). Peptide identifications were processed with Scaffold software. Confident protein identifications (probability > 0.9) with at least one unique peptide identifications and five MS/MS matches were considered. Detected variable gene family (if any) with most number of peptide and spectral matches is considered to be present in the deposit. The method was first validated in 8 cases of AL amyloidosis with known IGKV and IGLV region sequences and then applied on 1238 systemic and 393 localized AL cases. Differences between groups were measured by generating hypergeometric p-values.
In all 8 AL cases with known IGKV and IGLV region usage, gene family identified in the AL deposit via proteomics matched the gene family inferred from bone marrow plasma cells by Sanger sequencing of IGLC genes.
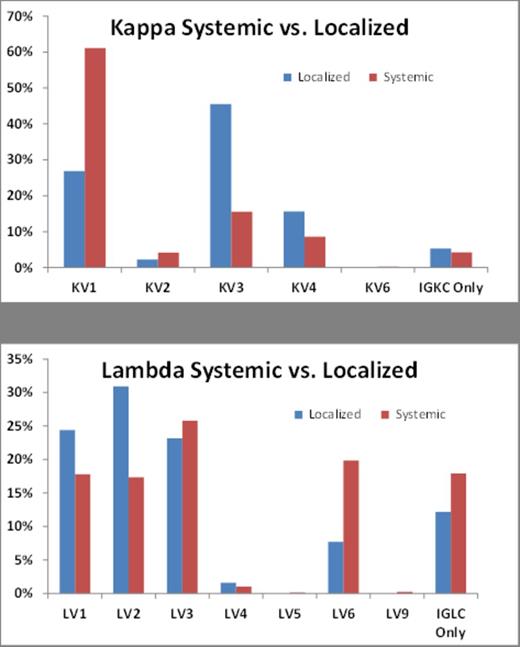
Armed with this method, we analyzed the amyloid proteomics data from 1238 patients with systemic AL amyloidosis and 393 patients with localized AL amyloidosis. The anatomical site distribution in systemic cohort was 296 GI tract, 364 heart, 225 kidney, 81 liver and 272 fat aspirate; Localized cohort has 78 bladder, 158 lung and 157 skin cases. Figure 1 shows the normalized frequency of the variable gene families detected between the systemic and localized AL amyloidosis. For AL-kappa, KV1 was more prevalent in systemic cases when compared to localized cases (p=7.2E-12) suggesting KV1 clones as a hallmark for systemic AL-kappa amyloidosis. KV3 was more frequently seen in localized AL-kappa cases when compared to systemic AL-kappa cases (p =2.8E-11). For AL-lambda, LV1 and LV2 gene families were more prevalent in localized cases when compared to systemic cases (p= 0.039 and 2.7E-05). LV6 was more prevalent in systemic AL-lambda cases (p=1.4E-07). We also detected a higher incidence of mixed AL/AH type in localized AL (18%) when compared to systemic AL (5%; Odds Ratio=4.2673, p<0.0001).
Frequency of Ig variable gene families observed in systemic and localized light chain amyloidosis (n=1617). “IGKC Only” and “IGLC Only” represent cases with no detectable variable region peptides.
Frequency of Ig variable gene families observed in systemic and localized light chain amyloidosis (n=1617). “IGKC Only” and “IGLC Only” represent cases with no detectable variable region peptides.
We next turned to the organ specific IGKV and IGLV gene family usage patterns in patients with localized amyloidosis. For 147 localized AL-kappa cases, KV3 was most dominant in lung (64%) and KV1 was dominant in skin (38%). For 246 localized AL-lambda cases, LV2 was most prevalent in lung (34%) and LV3 was most prevalent in skin (36%). For bladder, both LV1 and LV2 had comparable prevalence (42% and 32%).
For 361 systemic AL-kappa cases, KV1 consistently ranked at the top of gene families used in GI tract (55%), heart (71%), kidney (59%), liver (75%) and fat aspirate (50%). For 877 systemic AL-lambda cases, LV2 was predominant in GI tract (30%), LV3 in heart (35%) and LV6 in kidney (33%). The prevalence of LV1, LV2 and LV3 in liver is comparable (36%, 22% and 31%). LV6 and LV3 had comparable prevalence (22% and 24%) in AL-lambda fat aspirates.
The novel proteomics method detects IGLC V family usage in large cohorts of AL patients. It identifies unique profiles in systemic and localized cases, and in different organ sites. This information will be helpful in determining systemic versus localized nature of AL amyloidosis at diagnosis and to assess risk of specific end organ involvement. We found also a strong association between IGKV and IGLV gene usage and organ involvement.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.