Abstract
NOTCH1 is a proto-oncogene with activating mutations described in a variety of malignancies, including acute lymphoblastic leukemia (ALL), mantle cell lymphoma (MCL) and chronic lymphocytic leukemia (CLL). While the prognostic significance of NOTCH1 mutations remains controversial in ALL, recent data suggest that NOTCH1 PEST domain mutations are associated with adverse prognosis in patients with CLL. NOTCH1 mutations are found in around 8% of CLL patients at diagnosis, and since this disease has a heterogeneous clinical course and few prognostic markers, we aimed at designing a fast, cost effective and robust assay to detect NOTCH1 PEST domain mutations in patients with CLL. While 92% of the mutations in NOTCH1 PEST domain found in CLL are insertions or deletions, only 8% are represented by point mutations. Therefore we decided to use a fragment analysis approach in our assay. Given that a single mutation (c.7544_7545delCT), represents roughly 75% of all PEST domain mutations in CLL we designed a test that can, at the same time, detect the presence of this mutation specifically and also any insertion or deletion in exon 34.
We designed a PCR reaction using one FAM-labeled forward primer anchored at codon 2407 and two reverse primers. One specific for the c.7544_7545delCT mutation anchored at codon 2414 yielding a product of 356 base pairs (bp) and one anchored at codon 2425, yielding a product of 391 bp, comprising the hot spot for mutations in the NOTCH1 PEST domain. Primers were designed with Primer3 software (http://frodo.wi.mit.edu/) and the specificity of the reaction evaluated using the tool “PCR in silico” (http://genome.ucsc.edu/cgi-bin/hgPcr?command=start). The test yields three possible outputs:
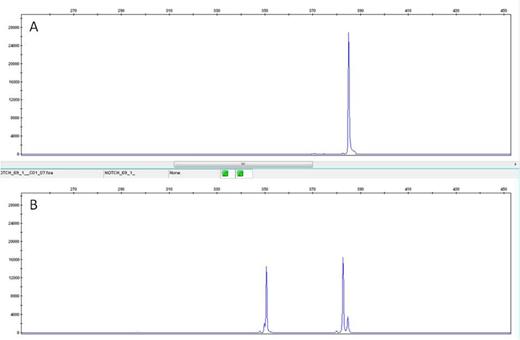
a) A single 391 bp peak: wild type samples
b) Three peaks (391 bp, 389 bp and 356 bp): heterozygous for c.7544_7545delCT
c) Two peaks (391 bp and another bigger or smaller, depending on the size of insertion / deletion): another insertion or deletion, but not c.7544_7545delCT.
We have studied 91 blood samples from CLL patients in diverse disease stages and found NOTCH1 mutations in 16 patients, (17.5%). When only patients with trisomy 12, isolated or with other cytogenetic abnormalities, were analyzed (N=16), we found an incidence of NOTCH1 mutations of 43% (7/16). At the same time, the frequency of NOTCH1 mutations in patients without trisomy 12 was 12% (9/75), in agreement with previous reports. All mutated cases (N=13) had the mutation c.7544_7545delCT.
In conclusion, we have designed a robust, fast and cost effective assay for routine identification of NOTCH1 PEST domain mutations, suitable for implementation in the clinical setting. Additionally we have found that the incidence and distribution among cytogenetic groups of NOTCH1 mutation in Brazilian patients with CLL is similar to the incidence described in other cohorts.
A – Wild Type NOTCH1 revealed by the presence of a single 391 bp peak.
B – Presence of heterozygous c.7544_7545delCT mutation evidenced by the presence of a 356 bp peak,corresponding to the allele specific pcr peak; and a double peak at 391 bp and 389 bp positions, corresponding to the wild type product (391 bp) and to the mutated product (389 bp) detected with the wild type primers.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.