Abstract
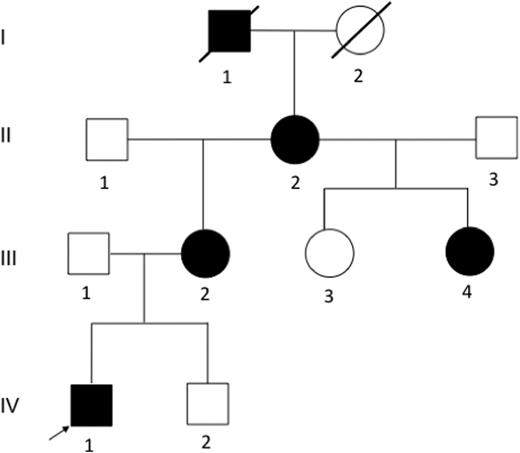
Immune thrombocytopenia (ITP) is an autoimmune-mediated bleeding disorder characterized by low blood platelet count. The etiology of ITP remains an area of active research, and the predisposing risk factors are unclear. Genetic studies on the hereditary form of ITP, which is very rare, may help to decipher the underlying cause of disease predisposition. Herein we report a four-generation family from Michigan with multiple members affected with ITP during childhood (Figure 1). The pedigree is consistent with an autosomal dominant inheritance pattern. We performed the first family-based genetic study of ITP using genome-wide linkage analysis in combination with whole genome sequencing (WGS) in order to identify the disease-causing mutation.
Pedigree of the Michigan family. Family members I1, II2, III2, III4, and IV1 were affected with primary ITP during childhood. DNA samples from II2, II3, III1, III2, III3, III4, IV1 and IV2 were available for this study.
Pedigree of the Michigan family. Family members I1, II2, III2, III4, and IV1 were affected with primary ITP during childhood. DNA samples from II2, II3, III1, III2, III3, III4, IV1 and IV2 were available for this study.
Peripheral blood specimens were collected from eight family members for DNA extraction and clinical information of the four affected individuals was obtained. Genome-wide genotyping was performed with high density Illumina HumanOmni1-Quad BeadChips, followed by linkage analysis with MERLIN. Two affected family members (II2 and IV1) were chosen for WGS on Illumina HiSeq with 30X coverage. Sequence alignment and variant calling were done with DNAnexus. The variants, which are common (minor allele frequency >=1%), outside of the linkage region, not shared by the two members, or silent mutations, were removed. Copy number variation (CNV) data obtained from the arrays were analyzed to identify co-segregation with the phenotype. Conservation of sequence, predicted impact on protein function and biological plausibility were considered to further filter the variants. The final disease-causing candidate variant NOS3 (nitric oxide synthase 3) c.1267G>A was validated by Sanger sequencing. The potential impact of this variant on the NOS3 protein function was evaluated by protein structure modeling. Furthermore, the wild-type and mutant NOS3 expression constructs were generated and transfected into COS-7 cells for transient protein expression. The total NOx production (nitrite and nitrate), measured with a HPLC assay, was normalized to expressed NOS3 and phospho-NOS3 protein, quantitated with western blot.
Mutations within the 5'UTR of ANKRD26 gene were reported to account for the autosomal dominant thrombocytopenia in some cases; however, no mutation was detected within the 5'UTR of ANKRD26 in this family and the linkage analysis also confirmed ANKRD26 gene being outside of the linkage region. The disease locus is mapped to a total of 215MB chromosomal region (6.92% of genome) by parametric linkage analysis with a LOD score of 1.1953. Within the linkage region, no CNV co-segregates with the phenotype. After removing common variants from those shared by II2 and IV1, no indel variants and 12 non-synonymous SNP variants were identified within the coding sequence. Following further removal of the SNP variants with low conservation scores or predicted by Polyphen to be a benign change on protein function, 5 variants from 5 genes remained, of which NOS3 is most biologically plausible based on the well-known role of NO in autoimmunity and oxidative stress. The co-segregation of the NOS3 variant with the phenotype of ITP in this family was validated via Sanger sequencing. Finally, an in vitro study demonstrated reduced NO production by mutant NOS3 protein compared to wild type NOS3 protein.
In this first genetic study of familial ITP, we identified the disease-causing mutation by using family-based linkage analysis and WGS. This finding will not only uncover the underlying genetic cause for the predisposition to childhood ITP in this Michigan kindred, but may also provide useful insights on the understanding of the pathogenesis of ITP in general.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.