Key Points
MLL-ENL targets long-term HSCs exclusively to develop leukemia in a novel conditional transgenic mouse through upregulation of Plzf.
Plzf is critically involved in the aberrant self-renewal program in HSCs induced by the MLL fusion gene.
Abstract
Oncogenic transformation requires unlimited self-renewal. Currently, it remains unclear whether a normal capacity for self-renewal is required for acquiring an aberrant self-renewal capacity. Our results in a new conditional transgenic mouse showed that a mixed lineage leukemia (MLL) fusion oncogene, MLL-ENL, at an endogenous-like expression level led to leukemic transformation selectively in a restricted subpopulation of hematopoietic stem cells (HSCs) through upregulation of promyelocytic leukemia zinc finger (Plzf). Interestingly, forced expression of Plzf itself immortalized HSCs and myeloid progenitors in vitro without upregulation of Hoxa9/Meis1, which are well-known targets of MLL fusion proteins, whereas its mutant lacking the BTB/POZ domain did not. In contrast, depletion of Plzf suppressed the MLL-fusion–induced leukemic transformation of HSCs in vitro and in vivo. Gene expression analyses of human clinical samples showed that a subtype of PLZF-high MLL-rearranged myeloid leukemia cells was closely associated with the gene expression signature of HSCs. These findings suggested that MLL fusion protein enhances the self-renewal potential of normal HSCs to develop leukemia, in part through a Plzf-driven self-renewal program.
Introduction
Malignant hematopoiesis in acute myeloid leukemia (AML) is thought to originate from a subpopulation of leukemia-initiating/leukemic stem cells (LICs/LSCs) with unlimited self-renewal, as in normal hematopoiesis organized hierarchically by hematopoietic stem cells (HSCs).1,2 Given the common properties, it has been proposed that in the process of leukemogenesis, LICs occur in committed progenitors acquiring aberrant self-renewal or in HSCs acquiring aberrant proliferation.3
Previous studies of mixed lineage leukemia (MLL) fusion oncogenes using AML models have unveiled the mechanisms of leukemogenesis, including the roles of downstream Hox and Meis1, the assembly of chromatin-modifying supercomplexes with histone modification and/or transcriptional elongation, and the biology of LICs.4-6 MLL fusion genes including MLL-ENL have been shown to confer an aberrant self-renewal capacity on myeloid-committed progenitors, leading to leukemic initiation.7,8 In contrast, constitutively active mutants of STAT5,9 which is activated by several oncogenes, are able to transform long-term HSCs (LT-HSCs defined as CD34–c-Kit+Sca-1+Lin– [KSL] cells) but not short-term HSCs (ST-HSCs defined as CD34+KSL cells).10,11 These contrasting findings led us to hypothesize that MLL fusion genes may preferentially transform ST-HSCs, rather than LT-HSCs. Interestingly, the molecular mechanism of leukemic transformation by MLL fusion genes has been shown to differ among the target cell subpopulations.12,13 Several studies have shown that MLL fusion genes can transform the entire population of KSL cells, including immature progenitors and HSCs,7,8 but no detailed studies have been reported on the hypothetical model.
The promyelocytic leukemia zinc finger (PLZF, also called ZBTB16) gene, which was originally identified as a partner gene fused with RARα in acute promyelocytic leukemia,14 encodes a transcription factor. PLZF recruits polycomb group proteins and histone deacetylases to target promoter regions, leading to gene repression,15 and it also activates gene expression through promoter binding.16 In hematopoiesis, PLZF is highly expressed in human stem and immature progenitor cells and is downregulated during myeloid differentiation.17-19 Studies using Plzf-deficient mice have shown that Plzf is essential for self-renewal in spermatogenesis, but not in hematopoiesis.20,21 However, the role of PLZF in leukemogenesis remains unclear, although PLZA-RARα has been extensively investigated.22
To clarify the molecular mechanism of leukemic transformation by MLL fusion genes, we developed a novel conditional MLL-ENL transgenic (Tg) mouse model. Unexpectedly, conditional expression of MLL-ENL, at a level comparable with that of endogenous Mll, selectively targeted LT-HSCs, but not ST-HSCs and myeloid progenitors (MPs), for leukemic transformation. We also found a previously unrecognized mechanism of MLL-fusion–mediated transformation in HSCs through a Plzf-driven self-renewal program.
Methods
Mice
A fragment containing the conditional FLAG-tagged MLL-ENL23 Tg cassette driven by hybrid cytomegalovirus (CMV) enhancer/chicken β-actin (CAG) promoter (Figure 1A) was injected into fertilized C57/BL6 (B6) mouse eggs, as previously described.24 All animal studies were approved by the Animal Care Committees of Mie University.
Leukemic transformation of Tg LT-HSCs conditionally expressing MLL-ENL. (A) Construction of the plasmid for conditional expression of MLL-ENL. CAG, hybrid CMV enhancer/chicken β-actin promoter; FLAG, tag sequence fused to the C terminus; SV40, simian virus 40. Arrowheads indicate loxP sites; filled diamond indicates triple stop codon sequence; horizontal arrows indicate primers to detect recombination between the loxP sites in (D); vertical arrow indicates the EcoR I site. (B) Expression of GFP in peripheral white blood cells from a conditional MLL-ENL Tg mouse (black line) compared with a WT littermate (gray shadow) on FACS. (C) Experimental strategy for myeloid immortalization assays and BMT. Sorted BM cells were retrovirally transduced with transgenes (dotted diagonal lines). (D) Recombination detected by genomic PCR in myeloid immortalization assays of WT and Tg LT-HSCs retrovirally transduced with mock (harvested at the end of the first plating) or CreER (Tg only; harvested at the end of each round of plating). G, germline configuration; R, recombination between loxP sites. (E) Expression levels of Hoxa9, Meis1, Evi1, and MLL-ENL by QRT-PCR in myeloid immortalization assays. Tg LT-HSCs (CD34[−]), Tg ST-HSCs (CD34[+]), Tg MPs, WT whole KSL cells, and WT MPs were retrovirally transduced with mock, CreER, or MLL-ENL and harvested at the first replating. (F) Myeloid immortalization assays of cells with retroviral transduction, as described in (E), by replating 104 cells. (G-H) Typical morphology of the colonies of the Tg LT-HSCs retrovirally transduced with CreER at the end of the third round (G), and the immortalized cells constituting the colonies (H). Colonies, and cells stained with Wright-Giemsa, were viewed with an Olympus CKX41 microscope using a 4×/0.13 objective lens and an Olympus BX41 microscope using a 20×/0.5 objective lens, respectively. Images were acquired with Olympus DP21 and Olympus DP21 software. Magnification and bars in (G) indicate 40× and 200 μm; magnification and bars in (H) indicate 200× and 20 μm. (I) Immunophenotype of the immortalized Tg LT-HSCs (CD34–KSL) and the WT whole KSL cells immortalized by retroviral transduction of MLL-ENL. (J) Survival curves of mice transplanted with Tg LT-HSCs (CD34–; n = 4), ST-HSCs (CD34+; n = 4), whole KSL cells (n = 5 for CreER; n = 3 for mock), and MPs (n = 4), retrovirally transduced with CreER(-IRES-EGFP) or mock. The bar graphs indicate the mean ± SD of 3 independent experiments.
Leukemic transformation of Tg LT-HSCs conditionally expressing MLL-ENL. (A) Construction of the plasmid for conditional expression of MLL-ENL. CAG, hybrid CMV enhancer/chicken β-actin promoter; FLAG, tag sequence fused to the C terminus; SV40, simian virus 40. Arrowheads indicate loxP sites; filled diamond indicates triple stop codon sequence; horizontal arrows indicate primers to detect recombination between the loxP sites in (D); vertical arrow indicates the EcoR I site. (B) Expression of GFP in peripheral white blood cells from a conditional MLL-ENL Tg mouse (black line) compared with a WT littermate (gray shadow) on FACS. (C) Experimental strategy for myeloid immortalization assays and BMT. Sorted BM cells were retrovirally transduced with transgenes (dotted diagonal lines). (D) Recombination detected by genomic PCR in myeloid immortalization assays of WT and Tg LT-HSCs retrovirally transduced with mock (harvested at the end of the first plating) or CreER (Tg only; harvested at the end of each round of plating). G, germline configuration; R, recombination between loxP sites. (E) Expression levels of Hoxa9, Meis1, Evi1, and MLL-ENL by QRT-PCR in myeloid immortalization assays. Tg LT-HSCs (CD34[−]), Tg ST-HSCs (CD34[+]), Tg MPs, WT whole KSL cells, and WT MPs were retrovirally transduced with mock, CreER, or MLL-ENL and harvested at the first replating. (F) Myeloid immortalization assays of cells with retroviral transduction, as described in (E), by replating 104 cells. (G-H) Typical morphology of the colonies of the Tg LT-HSCs retrovirally transduced with CreER at the end of the third round (G), and the immortalized cells constituting the colonies (H). Colonies, and cells stained with Wright-Giemsa, were viewed with an Olympus CKX41 microscope using a 4×/0.13 objective lens and an Olympus BX41 microscope using a 20×/0.5 objective lens, respectively. Images were acquired with Olympus DP21 and Olympus DP21 software. Magnification and bars in (G) indicate 40× and 200 μm; magnification and bars in (H) indicate 200× and 20 μm. (I) Immunophenotype of the immortalized Tg LT-HSCs (CD34–KSL) and the WT whole KSL cells immortalized by retroviral transduction of MLL-ENL. (J) Survival curves of mice transplanted with Tg LT-HSCs (CD34–; n = 4), ST-HSCs (CD34+; n = 4), whole KSL cells (n = 5 for CreER; n = 3 for mock), and MPs (n = 4), retrovirally transduced with CreER(-IRES-EGFP) or mock. The bar graphs indicate the mean ± SD of 3 independent experiments.
Retroviral constructs
CreER and MLL-ENL-ER,25 in which Cre and MLL-ENL were fused to the mutant ligand-binding domain of the estrogen receptor, respectively, MLL-ENL,23 MLL-SEPT6,23 E2A-HLF, Plzf, and Plzf mutants were cloned in a series of pMYs retroviral vectors.26 The pMYs-MLL-ENL-internal ribosomal entry site (IRES)-enhanced green fluorescent protein (EGFP) vector has been described elsewhere.27
Purification of mouse hematopoietic stem/progenitor cells
CD34–KSL, CD34+KSL, whole KSL, and c-Kit+Sca-1–Lin– (control fraction including MPs) cells were purified from bone marrow (BM) cells of 10- to 15-week-old hemizygous Tg or wild-type (WT) mice using fluorescence-activated cell sorting (FACS Aria; BD Biosciences) (supplemental Figure 1A, available on the Blood Web site).28
Retroviral transduction
Myeloid immortalization assay
Myeloid immortalization assays using serial replating were performed as previously described.23 Briefly, every 5 to 7 days, colonies were enumerated, followed by replating of the harvested cells (1 × 104) in the methylcellulose medium supplemented with stem cell factor, IL-3, IL-6, and granulocyte-macrophage colony-stimulating factor. The immortalized cells were harvested from colonies in the third plating.
FACS analysis
For intracellular flow cytometry of Plzf,30 cells were fixed and permeabilized using a Foxp3 Staining Buffer Set (eBioscience). Intracellular Plzf was detected by a mouse monoclonal anti-Plzf (D-9) antibody (Santa Cruz Biotechnology, Inc.), followed by staining with an allophycocyanin-conjugated anti-mouse IgG1 (Biolegend) with FACS Aria or Calibur (BD Biosciences).
Gene silencing by shRNA
shRNA sequences against Plzf, or luciferase as a control, were inserted into pMXsU6-KO (see supplemental Methods). The shRNA-transduced cells were sorted by Kusabira-Orange (KO) expression (top 10%-20%) with FACS Aria 72 hours after the transduction.
Transplantation
Leukemogenesis assays were performed as previously described.23,31 Briefly, for primary transplantation, MP (2 × 104) cells, whole KSL (2 × 103) cells, and cells arising from the respective 100 sorted cells in the Tg CD34– and CD34+ KSL populations retrovirally transduced with CreER and/or Plzf (supplemental Table 1) were transplanted into lethally irradiated (7.5 Gy) recipient B6 mice, together with 2 × 105 congenic BM cells, immediately after retroviral transduction. For secondary transplantation, splenocytes from leukemic mice receiving MLL-ENL-IRES-EGFP–transduced WT KSL cells were retrovirally transduced with shRNA/KO expressor, followed by sorting of GFP+ cells with high KO expression. The sorted cells (2 × 103) were immediately transplanted into sublethally irradiated (5.25 Gy) recipient B6 mice, or they were subjected to colony-forming assays. BM cells and splenocytes from morbid mice were analyzed as described elsewhere.23
Chromatin immunoprecipitation (ChIP)
Chromatin prepared from the immortalized Tg cells or MLL-ENL–immortalized WT MPs was precipitated using Dynabeads anti-Mouse IgG (Invitrogen) preincubated with a mouse monoclonal anti-FLAG (M2; Sigma-Aldrich) or a mouse IgG1 antibody (Biolegend) according to the manufacturer’s instruction. The purified DNA in precipitants was quantified by quantitative polymerase chain reaction.
Bioinformatics analysis
Gene set enrichment analyses (GSEAs) were performed using GSEA version 2.0 software (Broad Institute; http://www.broadinstitute.org/gsea) with Signal2Noise or Pearson metrics for gene ranking and 1000 data permutations.
Additional experimental methods are described in the supplemental Methods.
Results
Generation of conditional MLL-ENL Tg mice
We generated a conditional Tg construct including the EGFP cassette flanked by loxP sites and FLAG-tagged MLL-ENL (Figure 1A). Using this construct, DNA microinjections into 1200 fertilized mouse eggs were done, but only 1 of 3 obtained lines expressing GFP (Figure 1B) showed germline transmission that was also confirmed by Southern blot analysis (supplemental Figure 1B). Tg mice exhibited no hematologic abnormality until 15 months (data not shown).
Conditional expression of MLL-ENL selectively targets LT-HSCs for myeloid immortalization and induction of myeloproliferative disease-like leukemia
To investigate the immortalization potential of MLL-ENL in hematopoietic stem/progenitor subpopulations, Tg BM cells were sorted into previously well-defined fractionated cells CD34−KSL cells, CD34+KSL cells (hereafter referred to as LT-HSCs and ST-HSCs, respectively), and MPs (supplemental Figure 1A).10,32 These subpopulations of cells were retrovirally transduced with CreER (supplemental Figure 1C) and subjected to myeloid immortalization assays (Figure 1C). Unexpectedly, without 4-hydroxytamoxifen, recombination between loxP sites was detected in the CreER-transduced cells, even in the cells collected from the first-round colonies (hereafter referred to as the cells at the first replating) (Figure 1D), with expression of MLL-ENL (Figure 1E). With serial replating, the recombination bands were detected more clearly, whereas germline bands became more obscure (Figure 1D).
Surprisingly, only Tg LT-HSCs, and not Tg ST-HSCs and MPs, formed compact colonies comprising morphologically myelomonocytic blasts after serial replating, whereas initial plating of the CreER-transduced cells from the 3 subpopulations formed similar colonies with almost equal expression levels of MLL-ENL (Figure 1E-H; supplemental Figure 1D). WT KSL and MP cells retrovirally transduced with MLL-ENL showed replating ability.7 Immortalized Tg LT-HSCs exhibited characteristic immunophenotypes such as c-Kit+/Sca-1+, compared with retrovirally MLL-ENL-immortalized WT KSL cells (Figure 1I). We also confirmed the leukemogenic potential of the CreER-transduced cells using BM transplantation (BMT) assays (Figure 1J; supplemental Figure 1E). The majority of mice receiving the transduced whole KSL cells or LT-HSCs died of myeloproliferative disease–like leukemia with long latencies, whereas those receiving the transduced ST-HSCs or MPs did not.
Expression level of MLL-ENL transgene is comparable with that of endogenous murine Mll, but lower than that of retrovirally transduced MLL-ENL
To validate conditional expression of MLL-ENL, we examined its expression level in CreER–transduced cells at the first replating. MLL-ENL–transduced WT cells had a much higher expression of MLL-ENL than CreER-transduced Tg cells (Figure 1E); thus, expression of the MLL-ENL transgene was compared with that of the mouse endogenous Mll (Figure 2A and supplemental Figure 2A). The ratio of total MLL(-ENL)/Mll transcripts to Mll in mock and CreER-transduced cells was around 1 and 2, respectively, suggesting that expression levels of induced MLL-ENL were almost equal to those of Mll (Figure 2B-C). These results were confirmed by restriction fragment length polymorphism analyses (supplemental Figure 2B-D).
Characterization of Tg LT-HSCs conditionally expressing MLL-ENL. (A) Schematic representation of human MLL-ENL (MLL: thick; ENL: thin) transcript and the corresponding region of the endogenous transcript of mouse Mll. Mll and the total MLL-ENL/Mll transcripts were analyzed by QRT-PCR using common primers (horizontal arrows) with either Mll- or MLL/Mll-TaqMan probes. (B-C) Expression level of Mll (B) and relative ratio of total MLL(-ENL)/Mll to Mll transcripts (C), quantified by QRT-PCR. Tg LT-HSCs (CD34[−]), ST-HSCs (CD34[+]), and MPs retrovirally transduced with mock or CreER were harvested at the first replating in myeloid immortalization assays. (D) Expression levels of Hoxa9, Meis1, Evi1, and MLL-ENL by QRT-PCR in the immortalization assays. Tg LT-HSCs (CD34[−]) and ST-HSCs (CD34[+]) retrovirally transduced with CreER were harvested at the end of each round, and the first round, of plating, respectively. (E-F) Colony replating assays of subpopulations in the immortalized cells constituting colonies of CreER-transduced Tg LT-HSCs after the third round of plating, sorted on the basis of the immunophenotype (E). (G) Immunophenotype of the cells derived from the c-Kit+Sca-1+Gr-1– subpopulation of the immortalized Tg cells, harvested at the first replating. The bar graphs show the mean ± SD of 3 independent experiments.
Characterization of Tg LT-HSCs conditionally expressing MLL-ENL. (A) Schematic representation of human MLL-ENL (MLL: thick; ENL: thin) transcript and the corresponding region of the endogenous transcript of mouse Mll. Mll and the total MLL-ENL/Mll transcripts were analyzed by QRT-PCR using common primers (horizontal arrows) with either Mll- or MLL/Mll-TaqMan probes. (B-C) Expression level of Mll (B) and relative ratio of total MLL(-ENL)/Mll to Mll transcripts (C), quantified by QRT-PCR. Tg LT-HSCs (CD34[−]), ST-HSCs (CD34[+]), and MPs retrovirally transduced with mock or CreER were harvested at the first replating in myeloid immortalization assays. (D) Expression levels of Hoxa9, Meis1, Evi1, and MLL-ENL by QRT-PCR in the immortalization assays. Tg LT-HSCs (CD34[−]) and ST-HSCs (CD34[+]) retrovirally transduced with CreER were harvested at the end of each round, and the first round, of plating, respectively. (E-F) Colony replating assays of subpopulations in the immortalized cells constituting colonies of CreER-transduced Tg LT-HSCs after the third round of plating, sorted on the basis of the immunophenotype (E). (G) Immunophenotype of the cells derived from the c-Kit+Sca-1+Gr-1– subpopulation of the immortalized Tg cells, harvested at the first replating. The bar graphs show the mean ± SD of 3 independent experiments.
We next examined expression of genes regulated by MLL fusion genes in myeloid immortalization. At the first replating (Figure 1E), Hoxa9 and Meis1 were upregulated in the CreER-transduced cells, but were relatively weakly expressed, compared with the MLL-ENL–transduced cells. Evi1 was also upregulated in the CreER-transduced HSCs, particularly in the transduced LT-HSCs as highly as in the MLL-ENL–transduced KSL cells. With serial replating (Figure 2D), expression levels of the 3 genes in the CreER-transduced LT-HSCs were elevated. Taken together, these results demonstrate that our Tg mouse model resembles an Mll-fusion knock-in (KI) model rather than other mouse models using retroviral transduction of MLL fusion genes, suggesting that an endogenous-like level of MLL-ENL expression is sufficient for leukemic transformation in LT-HSCs, but not in ST-HSCs and MPs. However, because of heterogeneity of transgene expression among individual hematopoietic stem/progenitor cells in our model (Figure 1B and data not shown), we cannot formally exclude the possibility that the lack of transformation of MPs or ST-HSCs is caused by lack of MLL-ENL expression in the subset of cells with colony-forming or engraftment potential.
Immortalized Tg LT-HSCs can be subdivided into subpopulations with different self-renewal potentials
On the basis of FACS profiles of the immortalized LT-HSCs (Figure 1I), we examined whether expression of myeloid markers might be negatively correlated with the colony replating potential. Gr-1–, not Gr-1+, cells generated colonies similar to the parental cells, whereas CD11b– and CD11b+ cells generated the similar colonies (Figure 2E-F and data not shown). Interestingly, subfractionation of the Gr-1– cells by c-Kit/Sca-1 expression showed that only the c-Kit+Sca-1+Gr-1– subpopulation generated replatable colonies similar to the parental cells, whereas the other 2 subpopulation cells did not (Figure 2F-G).
MLL fusion genes enhance Plzf expression in HSCs
To investigate the molecular mechanism of leukemic transformation in LT-HSCs conditionally expressing MLL-ENL, we preliminarily performed comprehensive gene expression profiling of CreER-transduced LT-HSCs and ST-HSCs at the first replating using cDNA microarray analysis (GEO accession number: GSE48539), which revealed differential expression of Plzf (data not shown). Because Zbtb16 (Plzf) has been listed as a highly expressed gene in the comparison of Mll-AF9 KI with WT KSL cells,12 we investigated the expression levels of Plzf in the myeloid immortalization assays. At the first replating, Plzf expression was highest in LT-HSCs among the 3 subpopulations, and it was enhanced by induced MLL-ENL in LT/ST-HSCs, although these expression levels in the colony-forming cells were lower than that in freshly sorted (ie, pure) LT-HSCs (Figure 3A; supplemental Figure 3A-B). With serial replating, Plzf expression was further enhanced, leading to high expression in the Gr-1low/– subpopulation, including the Gr-1–/Sca-1+ subpopulation (Figure 3B). Plzf expression was also enhanced in the MLL-ENL– or MLL-SEPT6–transduced (Figure 1C; supplemental Figure 4) WT KSL, but not MP, cells at the first replating (Figure 3C). These results suggest that MLL fusion genes induced upregulation of Plzf specifically in HSCs, which might be associated with aberrant self-renewal activity in MLL-fusion–immortalized HSCs.
Myeloid immortalization of HSCs by MLL fusion genes associated with high Plzf expression. (A) Expression level of Plzf by QRT-PCR in myeloid immortalization assays. Tg LT-HSCs (CD34[−]), and ST-HSCs (CD34[+]) and MPs, retrovirally transduced with mock or CreER were harvested at the end of each round, and the first round, of plating, respectively. (B) Intracellular FACS analyses of Plzf expression in the immortalized cells constituting colonies of the CreER-transduced Tg LT-HSCs after the third round of plating. (C) Expression levels of Plzf by QRT-PCR in WT whole KSL or MP cells retrovirally transduced with mock, MLL-ENL, or MLL-SEPT6 at the first replating in myeloid immortalization assays. (D) Structure of Plzf (BTB/POZ domain and zinc fingers [Zn]) and a mutant (PlzfΔBTB) lacking the BTB/POZ domain, and expression of Plzf and the mutant by Western blot (lower left panels) and RT-PCR (lower right panels) analyses. Lysates extracted from Plat E cells transfected with mock (pMYs-IP), pMYs-Plzf-IP, or pMYs-PlzfΔBTB-IP were blotted with the anti-Plzf (αPlzf) antibody, followed by reprobe with the anti–α-tubulin antibody (αTub) as an internal control. The WT whole KSL and MP cells retrovirally transduced with mock, Plzf, or PlzfΔBTB were harvested at the first replating in myeloid immortalization assays and were subjected to RT-PCR. B2m was used as an internal standard. NTC, no template control. (E) Myeloid immortalization assays of WT LT-HSCs (CD34[−]), ST-HSCs (CD34[+]), whole KSL cells, and MPs retrovirally transduced with mock, Plzf, or PlzfΔBTB by replating 104 cells. (F-H) Typical morphology of the colonies (F) in the Plzf-transduced KSL cells at the end of the third round of plating, and typical morphology (G) and immunophenotype (H) of the cells constituting these colonies. Colonies, and cells stained with Wright-Giemsa, were viewed with an Olympus CKX41 microscope using a 4×/0.13 objective lens and an Olympus BX41 microscope using a 20×/0.5 objective lens, respectively. Magnification and bars in panel F indicate 40× and 200 μm; magnification and bars in (G) indicate 200× and 20 μm. (I) Expression levels of Hoxa9, Meis1, and Evi1 in the immortalization assays, compared with the CreER-transduced Tg LT-HSCs (CD34[−]) by QRT-PCR. WT whole KSL and MP cells were retrovirally transduced with mock or Plzf and harvested at the end of the first and third rounds of plating. The CreER-transduced Tg LT-HSCs were harvested at the end of the third round of plating. The bar graphs show the mean ± SD of 3 independent experiments.
Myeloid immortalization of HSCs by MLL fusion genes associated with high Plzf expression. (A) Expression level of Plzf by QRT-PCR in myeloid immortalization assays. Tg LT-HSCs (CD34[−]), and ST-HSCs (CD34[+]) and MPs, retrovirally transduced with mock or CreER were harvested at the end of each round, and the first round, of plating, respectively. (B) Intracellular FACS analyses of Plzf expression in the immortalized cells constituting colonies of the CreER-transduced Tg LT-HSCs after the third round of plating. (C) Expression levels of Plzf by QRT-PCR in WT whole KSL or MP cells retrovirally transduced with mock, MLL-ENL, or MLL-SEPT6 at the first replating in myeloid immortalization assays. (D) Structure of Plzf (BTB/POZ domain and zinc fingers [Zn]) and a mutant (PlzfΔBTB) lacking the BTB/POZ domain, and expression of Plzf and the mutant by Western blot (lower left panels) and RT-PCR (lower right panels) analyses. Lysates extracted from Plat E cells transfected with mock (pMYs-IP), pMYs-Plzf-IP, or pMYs-PlzfΔBTB-IP were blotted with the anti-Plzf (αPlzf) antibody, followed by reprobe with the anti–α-tubulin antibody (αTub) as an internal control. The WT whole KSL and MP cells retrovirally transduced with mock, Plzf, or PlzfΔBTB were harvested at the first replating in myeloid immortalization assays and were subjected to RT-PCR. B2m was used as an internal standard. NTC, no template control. (E) Myeloid immortalization assays of WT LT-HSCs (CD34[−]), ST-HSCs (CD34[+]), whole KSL cells, and MPs retrovirally transduced with mock, Plzf, or PlzfΔBTB by replating 104 cells. (F-H) Typical morphology of the colonies (F) in the Plzf-transduced KSL cells at the end of the third round of plating, and typical morphology (G) and immunophenotype (H) of the cells constituting these colonies. Colonies, and cells stained with Wright-Giemsa, were viewed with an Olympus CKX41 microscope using a 4×/0.13 objective lens and an Olympus BX41 microscope using a 20×/0.5 objective lens, respectively. Magnification and bars in panel F indicate 40× and 200 μm; magnification and bars in (G) indicate 200× and 20 μm. (I) Expression levels of Hoxa9, Meis1, and Evi1 in the immortalization assays, compared with the CreER-transduced Tg LT-HSCs (CD34[−]) by QRT-PCR. WT whole KSL and MP cells were retrovirally transduced with mock or Plzf and harvested at the end of the first and third rounds of plating. The CreER-transduced Tg LT-HSCs were harvested at the end of the third round of plating. The bar graphs show the mean ± SD of 3 independent experiments.
Plzf can drive an aberrant self-renewal program on HSCs and MPs
To examine whether Plzf itself may induce aberrant self-renewal activity, WT whole KSL cells, MPs, LT-HSCs, and ST-HSCs were retrovirally transduced with Plzf and subjected to myeloid immortalization assays and BMT assays (Figures 1C, 3D-E). All Plzf-transduced subpopulations, including ST-HSCs and MPs, were immortalized and generated similar compact colonies with myelomonocytic features at various differentiation stages, even in the sixth round of plating (Figure 3E-H and data not shown). In addition, Tg ST-HSCs and MPs, transduced with both CreER and Plzf, were also immortalized after serial replating (supplemental Figure 6A-B). However, neither the Plzf-transduced WT cells (KSL, MP) nor the doubly transduced Tg cells (ST-HSC, MP) led to lethal disease within 5 months, with no significant changes of peripheral blood (PB) cell counts (supplemental Table 1 and data not shown). Although almost all mice receiving the Plzf-transduced WT KSL cells displayed long-term engraftments of donor-derived cells in PB, none of the other groups of recipient mice did in PB (nor in BMs in some of the mice we examined) during the observational period (supplemental Figure 5 and data not shown). In immortalization by Plzf, expression levels of Hoxa9, Meis1, and Evi1, which were not significantly different between mock- and Plzf-transduced cells at the first replating, were mildly changed after serial replating. Notably, expression of the 3 genes after serial replating was much lower than that in the immortalized Tg LT-HSCs expressing relatively low levels of Plzf (Figure 3I and supplemental Figure 7). In contrast, a Plzf mutant lacking the BTB/POZ (broad-complex, tramtrack, and bric à brac/poxvirus and zinc finger)33 domain was incapable of immortalizing KSL and MP cells (Figure 3E). These results suggested that Plzf might drive an aberrant self-renewal program in HSCs and progenitors without upregulation of Hoxa9, Meis1, and Evi1, whereas other molecules are required for leukemic transformation of Tg ST-HSCs and MPs by MLL-ENL in vivo.
Plzf is critical for MLL-fusion–mediated immortalization of HSCs
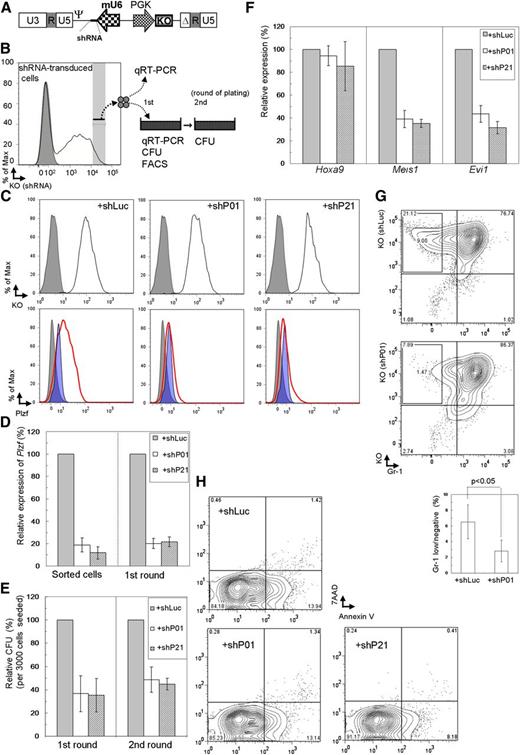
To further examine the involvement of Plzf in the immortalized Tg LT-HSCs, Plzf was depleted in the immortalized cells using shRNA (Figure 4A-B). Two shRNA sequences, shP01 and shP21, showed almost equivalent effects of depleting Plzf in the MLL-ENL–immortalized WT KSL cells that had been retrovirally transduced with Plzf (Figure 4C), and in the immortalized LT-HSCs (Figure 4D). Plzf knockdown reduced clonogenicity to about 40% of that in controls (Figure 4E), accompanied with decreased expression of Meis1 and Evi1, but not Hoxa9 (Figure 4F). Interestingly, reduction of clonogenicity was significantly correlated with reduction of the Gr-1low/– subpopulation with high potential for colony replating (Figure 4G), whereas it was not correlated with apoptosis (Figure 4H).
Suppressive effects by Plzf depletion on clonogenicity of Tg LT-HSCs immortalized by conditional expression of MLL-ENL. (A) Structure of the retroviral vector, pMXsU6-KO, carrying the expression cassette of shRNA (short thick line) driven by the mouse U6 (mU6) promoter and KO driven by the PGK promoter in the self-inactivated backbone. Δ, deletion of a fragment containing enhancer elements in the U3 sequence; Ψ, packaging signal. (B) Experimental strategy using immortalized cells with depletion of Plzf by retroviral transduction of shRNA and KO coexpressor. (C) Evaluation of Plzf depletion by shRNA (shP01 or shP21) using the MLL-ENL–immortalized WT KSL (ME) cells. ME cells were retrovirally transduced with Plzf (ME+Plzf) or mock (ME+IP) followed by drug selection. The shRNA-transduced ME+Plzf cells at the first replating were assessed on intracellular FACS. Cells transduced with the shRNA expressor confirmed by KO expression (black line) are shown in the upper panels, where gray shadows represent parental ME+Plzf cells. Plzf expression in shRNA-transduced ME+Plzf and ME+IP cells (as references) is depicted with red thick lines and blue shadow profiles, respectively, in the lower panels, where the gray shadows represent staining with the isotype control antibody in shRNA-transduced ME+Plzf cells. (D-E) Expression levels of Plzf by QRT-PCR (D) and relative colony-forming units (CFU) (E) of the cells sorted from the shRNA-transduced immortalized Tg LT-HSCs in colony replating assays. (F-H) Expression levels of Hoxa9, Meis1, and Evi1 by QRT-PCR (F) and quantification of Gr-1low/– (G) and apoptotic (H) subpopulations by FACS in the Plzf-depleted immortalized LT-HSCs at the first replating. shLuc, shRNA against luciferase; shP01 and shP21, different shRNAs against Plzf. The bar graphs show the mean ± SD of 3 independent experiments.
Suppressive effects by Plzf depletion on clonogenicity of Tg LT-HSCs immortalized by conditional expression of MLL-ENL. (A) Structure of the retroviral vector, pMXsU6-KO, carrying the expression cassette of shRNA (short thick line) driven by the mouse U6 (mU6) promoter and KO driven by the PGK promoter in the self-inactivated backbone. Δ, deletion of a fragment containing enhancer elements in the U3 sequence; Ψ, packaging signal. (B) Experimental strategy using immortalized cells with depletion of Plzf by retroviral transduction of shRNA and KO coexpressor. (C) Evaluation of Plzf depletion by shRNA (shP01 or shP21) using the MLL-ENL–immortalized WT KSL (ME) cells. ME cells were retrovirally transduced with Plzf (ME+Plzf) or mock (ME+IP) followed by drug selection. The shRNA-transduced ME+Plzf cells at the first replating were assessed on intracellular FACS. Cells transduced with the shRNA expressor confirmed by KO expression (black line) are shown in the upper panels, where gray shadows represent parental ME+Plzf cells. Plzf expression in shRNA-transduced ME+Plzf and ME+IP cells (as references) is depicted with red thick lines and blue shadow profiles, respectively, in the lower panels, where the gray shadows represent staining with the isotype control antibody in shRNA-transduced ME+Plzf cells. (D-E) Expression levels of Plzf by QRT-PCR (D) and relative colony-forming units (CFU) (E) of the cells sorted from the shRNA-transduced immortalized Tg LT-HSCs in colony replating assays. (F-H) Expression levels of Hoxa9, Meis1, and Evi1 by QRT-PCR (F) and quantification of Gr-1low/– (G) and apoptotic (H) subpopulations by FACS in the Plzf-depleted immortalized LT-HSCs at the first replating. shLuc, shRNA against luciferase; shP01 and shP21, different shRNAs against Plzf. The bar graphs show the mean ± SD of 3 independent experiments.
To investigate the Plzf-mediated self-renewal program in myeloid immortalization by MLL fusion genes more generally, Plzf was depleted in WT KSL cells retrovirally immortalized by MLL-ENL, MLL-SEPT6, and E2A-HLF (supplemental Figure 8A) that functions in a different manner from MLL fusion genes25 and MLL-ENL-immortalized WT MP cells (Figure 1C). Similar levels of Plzf depletion (Figure 5A) reduced clonogenicity to about 50% to 60% of that in controls in MLL-ENL– and MLL-SEPT6–immortalized KSL cells, in contrast to little reduction of clonogenicity in E2A-HLF–immortalized KSL and MLL-ENL–immortalized MP cells (Figure 5A and supplemental Figure 8B). We also examined restoration of the reduced clonogenicity by shP01-resistant Plzf expression (encoded by Plzf*) (Figure 5B), using the MLL-ENL–immortalized KSL cells that were retrovirally transduced with Plzf* or its mutant lacking the BTB/POZ domain (Plzf*/ΔBTB) (Figure 5C and supplemental Figure 8C). Quantitative reverse transcription-polymerase chain reaction (QRT-PCR) analyses in the coding region (Figure 5B) revealed little reduction of overall Plzf expression, whereas those in the 3′ untranslated region revealed an effective reduction of endogenous Plzf expression (Figure 5D). The reduction of clonogenicity by shP01 was almost completely overcome by Plzf*, but was hardly at all by Plzf*/ΔBTB (Figure 5E; supplemental Figure 8D).
Rescue of MLL-fusion–immortalized KSL cells from Plzf depletion by transduction with shRNA-resistant Plzf, but not with a resistant Plzf lacking the BTB/POZ domain. (A) Expression levels of Plzf by QRT-PCR (upper) and relative CFU (lower) of the cells sorted from shRNA-transduced immortalized cells in colony forming assays. WT whole KSL and MP cells immortalized by retroviral transduction of MLL-ENL, MLL-SEPT6, or E2A-HLF were harvested at the end of the first plating. (B) Structures of Plzf and a mutant lacking the BTB/POZ domain, modified with introduction of silent mutations (*) resistant to shRNA (shP01) against Plzf. Arrows indicate primers to detect Plzf transcripts in the coding region. (C) Expression levels of the mutated Plzf by QRT-PCR in the MLL-ENL–immortalized WT KSL (ME) cells retrovirally transduced with Plzf* (ME+Plzf*) or Plzf*/ΔBTB (ME+Plzf*/ΔBTB). (D-E) Expression levels of Plzf by QRT-PCR (D) and relative CFU (E) of the cells sorted from shRNA-transduced ME+Plzf* or ME+Plzf*/ΔBTB cells in colony replating assays. QRT-PCR analyses were performed using primers in the coding region (A [upper], C,D [upper]) or the 3′ untranslated region (D [lower]). The bar graphs show the mean ± SD of 3 independent experiments.
Rescue of MLL-fusion–immortalized KSL cells from Plzf depletion by transduction with shRNA-resistant Plzf, but not with a resistant Plzf lacking the BTB/POZ domain. (A) Expression levels of Plzf by QRT-PCR (upper) and relative CFU (lower) of the cells sorted from shRNA-transduced immortalized cells in colony forming assays. WT whole KSL and MP cells immortalized by retroviral transduction of MLL-ENL, MLL-SEPT6, or E2A-HLF were harvested at the end of the first plating. (B) Structures of Plzf and a mutant lacking the BTB/POZ domain, modified with introduction of silent mutations (*) resistant to shRNA (shP01) against Plzf. Arrows indicate primers to detect Plzf transcripts in the coding region. (C) Expression levels of the mutated Plzf by QRT-PCR in the MLL-ENL–immortalized WT KSL (ME) cells retrovirally transduced with Plzf* (ME+Plzf*) or Plzf*/ΔBTB (ME+Plzf*/ΔBTB). (D-E) Expression levels of Plzf by QRT-PCR (D) and relative CFU (E) of the cells sorted from shRNA-transduced ME+Plzf* or ME+Plzf*/ΔBTB cells in colony replating assays. QRT-PCR analyses were performed using primers in the coding region (A [upper], C,D [upper]) or the 3′ untranslated region (D [lower]). The bar graphs show the mean ± SD of 3 independent experiments.
Taken together, these results suggest that Plzf plays a critical role in the aberrant self-renewal activity conferred by MLL fusion genes in HSCs.
Plzf is also critical for MLL-fusion–induced leukemogenesis of HSCs in vivo
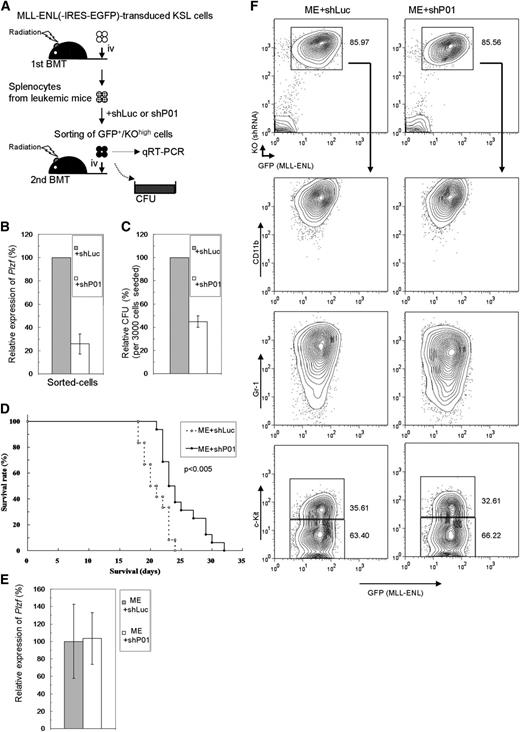
Involvement of Plzf in leukemogenesis of HSCs by MLL-ENL was next assessed in vivo. Primary leukemic cells derived from MLL-ENL–transduced WT KSL cells were retrovirally transduced with shRNA/KO expressor, and cells highly expressing KO were secondarily transplanted or subjected to colony-forming assays (Figure 6A). In the sorted cells, Plzf expression was effectively depleted by shP01 (Figure 6B), and clonogenicity was reduced to about 40% of that in controls (Figure 6C). These Plzf-depleted cells developed leukemia with a significantly prolonged latency (Figure 6D), without morphologic and immunophenotypical differences (Figure 6F and data not shown). There were no significant differences in Plzf and KO expression in the secondarily leukemic cells (Figure 6E-F), implying enrichment of less effectively Plzf-depleted cells during the course of the disease progression. These results suggest that Plzf was also critical for aberrant self-renewal activity in HSCs transformed by MLL fusion genes in vivo.
Plzf depletion prolongs MLL-ENL–induced leukemia. (A) Experimental strategy for the leukemia model using secondary BMT of primary MLL-ENL–leukemic cells. (B-C) Expression levels of Plzf by QRT-PCR (B) and relative CFU (C) of the cells sorted from shRNA-transduced leukemic cells. (D) Survival curves of the mice secondarily transplanted with the same Plzf-depleted (ME+shP01; n = 16) or control (ME+shLuc; n = 12) leukemic cells that are analyzed in panels B,C. Data from 3 independent experiments using independent primary leukemic cells were combined. (E-F) Expression levels of Plzf by QRT-PCR (E) and typical immunophenotype (F) of splenocytes from disease mice. The bar graphs show the mean ± SD of 3 independent experiments.
Plzf depletion prolongs MLL-ENL–induced leukemia. (A) Experimental strategy for the leukemia model using secondary BMT of primary MLL-ENL–leukemic cells. (B-C) Expression levels of Plzf by QRT-PCR (B) and relative CFU (C) of the cells sorted from shRNA-transduced leukemic cells. (D) Survival curves of the mice secondarily transplanted with the same Plzf-depleted (ME+shP01; n = 16) or control (ME+shLuc; n = 12) leukemic cells that are analyzed in panels B,C. Data from 3 independent experiments using independent primary leukemic cells were combined. (E-F) Expression levels of Plzf by QRT-PCR (E) and typical immunophenotype (F) of splenocytes from disease mice. The bar graphs show the mean ± SD of 3 independent experiments.
Upregulation of PLZF in MLL-rearranged AMLs is correlated with HSC expression signature
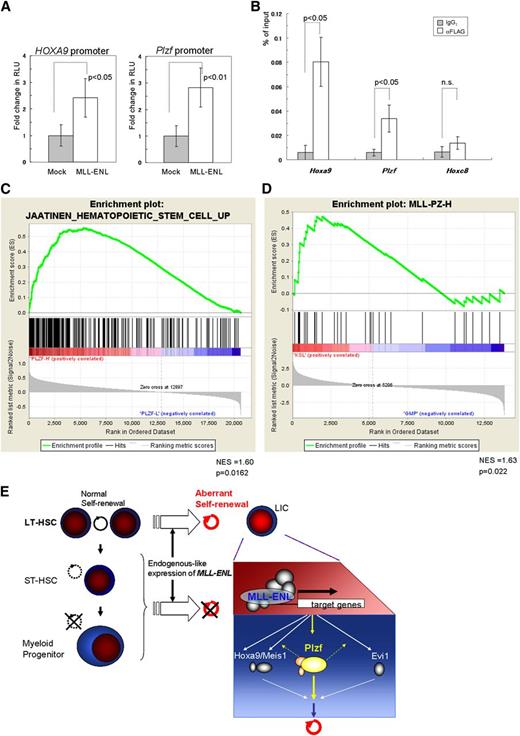
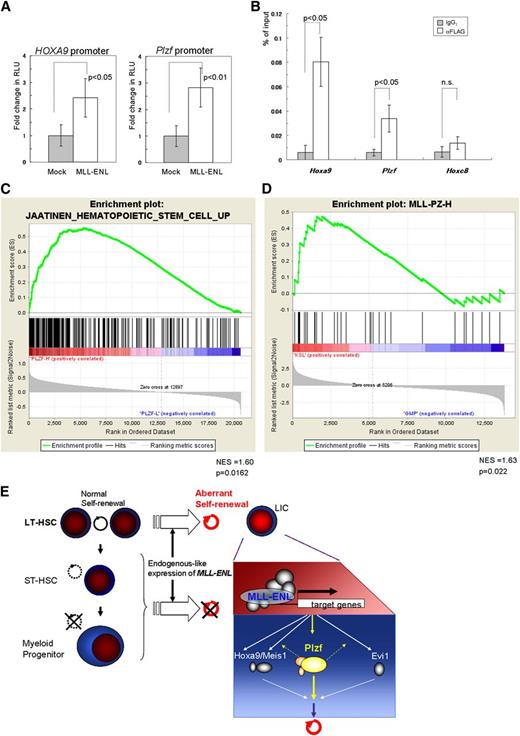
We finally investigated the molecular mechanism of upregulation of Plzf in leukemic transformation of HSCs by MLL-ENL using reporter and ChIP assays. Luciferase assays in K562 cells revealed that MLL-ENL activated the reporter gene through the Plzf promoter region and through the HOXA9 promoter region (Figure 7A). ChIP analyses showed a significant increase of MLL-ENL binding to the Plzf promoter in the immortalized Tg LT-HSCs (Figure 7B), but not in the MLL-ENL–immortalized WT MPs (supplemental Figure 9A). In addition, in inducibly MLL-ENL-ER25 –immortalized WT KSL cells, inactivation of MLL-ENL-ER by withdrawal of 4-hydroxytamoxifen reduced Plzf expression substantially in a time course–dependent manner, before suppression of cellular proliferation at day 3 (supplemental Figure 9B-C). These results suggest that MLL-ENL drives aberrant expression of Plzf in HSCs.
Close association between the MLL fusion gene and Plzf/PLZF expression in HSCs. (A) Reporter assays using luciferase driven by the mouse Plzf and human HOXA9 promoters, respectively. RLU, relative light units. (B) Relative binding of MLL-ENL (detected by an anti-FLAG antibody) to Hoxa9, Plzf, and Hoxc8 (used as a negative control) promoter regions in the Tg LT-HSCs immortalized by conditional expression of FLAG-tagged MLL-ENL. (C) GSEA of MLL-rearranged AML samples (GSE17855) showing that a gene set upregulated in HSCs35 was enriched in clinical samples expressing high levels of PLZF compared with those expressing low levels of PLZF. (D) GSEA of normal mouse KSL and GMP cells (GSE10627) showing that a gene set (MLL-PZ-H) representing the MLL-rearranged AML samples with high PLZF expression was enriched in KSL cells compared with GMP cells. (C-D) GSEAs were performed with Signal2Noise metric. NES, normalized enrichment score. (E) A proposed model of the leukemogenesis by endogenous-like expression of MLL-ENL. In contrast to retroviral transduction leading to strong expression, relatively weak but endogenous-like expression of MLL-ENL selectively transforms LT-HSCs into leukemic initiating cells (LICs), at least partly through upregulation of Plzf. The bar graphs show the mean ± SD of 3 independent experiments.
Close association between the MLL fusion gene and Plzf/PLZF expression in HSCs. (A) Reporter assays using luciferase driven by the mouse Plzf and human HOXA9 promoters, respectively. RLU, relative light units. (B) Relative binding of MLL-ENL (detected by an anti-FLAG antibody) to Hoxa9, Plzf, and Hoxc8 (used as a negative control) promoter regions in the Tg LT-HSCs immortalized by conditional expression of FLAG-tagged MLL-ENL. (C) GSEA of MLL-rearranged AML samples (GSE17855) showing that a gene set upregulated in HSCs35 was enriched in clinical samples expressing high levels of PLZF compared with those expressing low levels of PLZF. (D) GSEA of normal mouse KSL and GMP cells (GSE10627) showing that a gene set (MLL-PZ-H) representing the MLL-rearranged AML samples with high PLZF expression was enriched in KSL cells compared with GMP cells. (C-D) GSEAs were performed with Signal2Noise metric. NES, normalized enrichment score. (E) A proposed model of the leukemogenesis by endogenous-like expression of MLL-ENL. In contrast to retroviral transduction leading to strong expression, relatively weak but endogenous-like expression of MLL-ENL selectively transforms LT-HSCs into leukemic initiating cells (LICs), at least partly through upregulation of Plzf. The bar graphs show the mean ± SD of 3 independent experiments.
To examine involvement of PLZF in MLL-rearranged AML, we performed GSEAs of an MLL-rearranged AML cohort (GSE17855)34 using the top (PLZFhigh) and bottom (PLZFlow) 25th percentile samples subdivided on the basis of PLZF expression. These results showed positive enrichment of HSC-associated upregulated genes35 in the PLZFhigh group and negative (but insignificant) enrichment of HSC-associated downregulated genes (Figure 7C and data not shown). The same enrichment was also shown in another cohort (GSE1957736 ; supplemental Figure 9D). Expression data in mouse normal KSL and GMP cells (GSE10627)12 were also analyzed with GSEAs using 2 gene sets (supplemental Table 2) representing the PLZFhigh and PLZFlow groups, respectively, and showed positive enrichment of the PLZFhigh gene set in the KSL cells (Figure 7D). Interestingly, the PLZFhigh gene set included Gata2 and Dnmt3b, which are critical for self-renewal of HSCs,37,38 and both genes were more highly expressed in the immortalized Tg LT-HSCs with high expression of Plzf, compared with the MLL-ENL–immortalized WT MPs with low Plzf expression (supplemental Figure 9E). These results suggest that a subgroup of human MLL-rearranged AMLs with high PLZF expression has features characteristic of HSCs. Taken together, the results indicate that MLL fusion genes in human HSCs may induce AML, at least partly through upregulation of PLZF that drives an aberrant self-renewal program.
Discussion
The present study shows that LT-HSCs are exclusively targeted for leukemic transformation by endogenous-like expression of MLL-ENL (Figure 7E), which is compatible with a previous study12 showing that Mll-AF9-KI KSL cells are the most susceptible to the transformation. However, we also unveiled a striking difference in the susceptibility between LT-HSCs and ST-HSCs within whole KSL cells. Common lymphoid progenitors, which also lead to leukemic transformation in Mll-AF9 KI mice,12 were not immortalized by conditional expression of MLL-ENL (data not shown), presumably because of the difference of MLL-fusions and/or the difference in the timing of induction of gene expression. Our finding regarding MPs is in sharp contrast to previous studies7,8 showing that MLL fusion genes strongly driven by a retroviral promoter are able to transform MPs. Endogenous-like expression of an MLL fusion gene may enhance the self-renewal potential in LT-HSCs by hijacking the normal self-renewal program through elevated Plzf (Figure 7E), which has been obscured in previous studies using the whole population of KSL cells.7,8,12
Our conditional Tg mice model showed that MLL-ENL elevates Plzf expression in HSCs, which was much higher in LT-HSCs than in ST-HSCs, in accordance with the clear difference in leukemic transformation. MLL-SEPT6 also elevated Plzf expression in the whole population of KSL cells. The absence of elevation of Plzf in the MLL-ENL–immortalized WT MPs is consistent with the lack of an effect of Plzf depletion in these cells, suggesting that overexpression of MLL fusion genes may transform MPs through Plzf-independent pathways. Array data from Mll-AF9 KI mice showed Plzf expression in KI CMP/GMP cells, as in KI KSL cells, and higher expression in KI CMP/GMP cells than in WT CMP/GMP cells, presumably because of the same reasons as discussed before. Interestingly, Plzf was highly expressed in the more replatable Gr-1low/– subpopulations of the immortalized Tg LT-HSCs, and its overexpression immortalized WT LT-HSCs, ST-HSCs, and MPs. Plzf has important roles in cell fate determination,19,39 and PLZF overexpression induces propagation of human undifferentiated progenitor cells,18 supporting our result regarding the immortalization. The current study also suggests a critical function of the BTB/POZ domain in Plzf-mediated immortalization, although further characterization of this finding is needed.
Plzf depletion suppressed clonogenicity of the MLL-fusion–immortalized HSCs, accompanied with reduction of the Gr-1low/– subpopulations with replatable potential, suggesting the involvement of Plzf in the aberrant self-renewal capacity of MLL-fusion–immortalized HSCs. In contrast, Plzf depletion had little effect on KSL cells immortalized by E2A-HLF, a Bmi-1–independent oncogene.40 Complementary experiments in the depleted cells using Plzf or its mutant highlighted the requirement of the BTB/POZ domain in the Plzf-mediated immortalization. The BTB/POZ domain has an important role in the function of Plzf itself, through its interaction with various molecules including Bmi-1,33 whereas another study41 showed that MLL-AF9 immortalizes Bmi-1−/− c-Kit+BM cells. Thus, interaction of Plzf with associated molecules through the BTB/POZ domain might be important for MLL-fusion–mediated immortalization of HSCs.
Plzf depletion in AML arising from MLL-ENL–transduced KSL cells also prolonged latencies of the disease in secondary BMT, reminiscent of recent findings for Evi1.13,42 Conditional deletion of Evi1 in the MLL-ENL–immortalized KSL cells reduced clonogenicity,13 and Evi1 depletion in the Mll-AF9-KI leukemic cell line prolonged the latency of leukemia in BMT.42 Interestingly, our results demonstrated that the Plzf-immortalized cells expressed lower levels of Hoxa9, Meis1, and Evi1 than the immortalized Tg LT-HSCs, whereas Plzf depletion in the immortalized LT-HSCs reduced the expression levels of Meis1 and Evi1. Taken together with the results from the reporter and ChIP assays, and inactivation experiments of MLL-ENL-ER, this suggests that MLL fusion genes may transform HSCs at least partly through upregulation of Plzf by bypassing the Hoxa9/Meis1-dependent pathway, whereas Plzf may be involved in maintenance of the expression of Meis1 and Evi1 in the transformation (Figure 7E). Furthermore, in support of our mouse model, GSEAs suggested that MLL-rearranged AML with high expression of PLZF is closely associated with the properties of HSCs, similar to MLL-rearranged AML highly expressing Evi-1.13 Interestingly, GSEAs of a non–MLL-rearranged AML cohort (GSE17855)34 also showed enrichment of HSC-associated upregulated genes35 in PLZF-high samples (data not shown), implying the involvement of PLZF in non–MLL-rearranged AML. Further studies are needed to define the individual or interactive roles of these molecules, including Plzf, in the transformation of HSCs.
In conclusion, our study shows that LT-HSCs were selectively targeted for leukemic transformation by MLL-ENL, which was conditionally expressed at an endogenous-like level. This study also demonstrated the critical role of Plzf in MLL-fusion–mediated transformation of HSCs in vitro and in vivo. A subtype of MLL-rearranged AML highly expressing PLZF was closely associated with the properties of HSCs in gene expression signature. Because Plzf itself is not essential for normal hematopoiesis, the Plzf-driven aberrant program in leukemic transformation may be a target for molecular therapy against MLL-rearranged AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked ‘‘advertisement’’ in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Tsutomu Kamisako and Mamoru Ito for generating Tg mice, Atsushi Iwama for important technical advice, Nobuyuki Yajima for pCAG-lox-GFP, Christopher Baum for the plasmid containing Cre, Katsutoshi Ozaki for the construct containing the HOXA9 promoter, Hiroyuki Miyoshi for pCMV-VSV-G, and PALABRA Inc. (Kyoto, Japan) for language assistance.
This work was supported in part by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology in Japan; the Japan Leukaemia Research Fund (T.N., R.O.); Chugai Pharmaceutical Company Ltd, the Novartis Foundation for the Promotion of Science, and the Association for Research on Lactic Acid Bacteria (T.N.).
Authorship
Contribution: R.O. and T.N. designed the research; R.O., M.M., H.N., Y.E., E.M., A.N., S.I., K.S., and F.S.-M. performed experiments; R.O., M.M., N.K., T.K., and T.N. analyzed the results; and R.O. and T.N. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for Y.E. is the Laboratory of Cell Growth and Differentiation, Institute of Molecular and Cellular Biosciences, The University of Tokyo, Bunkyo-ku, Tokyo, Japan.
Correspondence: Tetsuya Nosaka, Department of Microbiology and Molecular Genetics, Mie University Graduate School of Medicine, 2-174 Edobashi, Tsu 514-8507, Japan; e-mail: nosaka@doc.medic.mie-u.ac.jp.

![Figure 1. Leukemic transformation of Tg LT-HSCs conditionally expressing MLL-ENL. (A) Construction of the plasmid for conditional expression of MLL-ENL. CAG, hybrid CMV enhancer/chicken β-actin promoter; FLAG, tag sequence fused to the C terminus; SV40, simian virus 40. Arrowheads indicate loxP sites; filled diamond indicates triple stop codon sequence; horizontal arrows indicate primers to detect recombination between the loxP sites in (D); vertical arrow indicates the EcoR I site. (B) Expression of GFP in peripheral white blood cells from a conditional MLL-ENL Tg mouse (black line) compared with a WT littermate (gray shadow) on FACS. (C) Experimental strategy for myeloid immortalization assays and BMT. Sorted BM cells were retrovirally transduced with transgenes (dotted diagonal lines). (D) Recombination detected by genomic PCR in myeloid immortalization assays of WT and Tg LT-HSCs retrovirally transduced with mock (harvested at the end of the first plating) or CreER (Tg only; harvested at the end of each round of plating). G, germline configuration; R, recombination between loxP sites. (E) Expression levels of Hoxa9, Meis1, Evi1, and MLL-ENL by QRT-PCR in myeloid immortalization assays. Tg LT-HSCs (CD34[−]), Tg ST-HSCs (CD34[+]), Tg MPs, WT whole KSL cells, and WT MPs were retrovirally transduced with mock, CreER, or MLL-ENL and harvested at the first replating. (F) Myeloid immortalization assays of cells with retroviral transduction, as described in (E), by replating 104 cells. (G-H) Typical morphology of the colonies of the Tg LT-HSCs retrovirally transduced with CreER at the end of the third round (G), and the immortalized cells constituting the colonies (H). Colonies, and cells stained with Wright-Giemsa, were viewed with an Olympus CKX41 microscope using a 4×/0.13 objective lens and an Olympus BX41 microscope using a 20×/0.5 objective lens, respectively. Images were acquired with Olympus DP21 and Olympus DP21 software. Magnification and bars in (G) indicate 40× and 200 μm; magnification and bars in (H) indicate 200× and 20 μm. (I) Immunophenotype of the immortalized Tg LT-HSCs (CD34–KSL) and the WT whole KSL cells immortalized by retroviral transduction of MLL-ENL. (J) Survival curves of mice transplanted with Tg LT-HSCs (CD34–; n = 4), ST-HSCs (CD34+; n = 4), whole KSL cells (n = 5 for CreER; n = 3 for mock), and MPs (n = 4), retrovirally transduced with CreER(-IRES-EGFP) or mock. The bar graphs indicate the mean ± SD of 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/7/10.1182_blood-2012-09-456665/4/m_1271f1.jpeg?Expires=1768288610&Signature=BbGlxXLGR7zj2tJV2B4g30yRf~aX~6d8w9qwiTbSP6fgkBg4Lj9cFGNTw-4qgxZb3jY8RYjZNN398XRdVgLARr8ij0-PqPYlypZbW09IWAvUQkpiOIEo1I3PLpQelr~K21y~YDV84TcU3lPF5cL5p2wqZZg8h8UlpcDsgxV~plB3EwQ8TMWR71U8glujyfMdSoaaOg-OUUUm9wCKfoL5h2mCX5yyptA0jWcNUm598dcEg2pp5xiaNFNPWb9EqeH1WpD2pwae8j7JDfHZtwJmvHS3dMMoGmh86rFtDM1LnGzrObMzON68Ey20toiNK13HVJxuHMXA3HICz-Z1SPnisw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. Characterization of Tg LT-HSCs conditionally expressing MLL-ENL. (A) Schematic representation of human MLL-ENL (MLL: thick; ENL: thin) transcript and the corresponding region of the endogenous transcript of mouse Mll. Mll and the total MLL-ENL/Mll transcripts were analyzed by QRT-PCR using common primers (horizontal arrows) with either Mll- or MLL/Mll-TaqMan probes. (B-C) Expression level of Mll (B) and relative ratio of total MLL(-ENL)/Mll to Mll transcripts (C), quantified by QRT-PCR. Tg LT-HSCs (CD34[−]), ST-HSCs (CD34[+]), and MPs retrovirally transduced with mock or CreER were harvested at the first replating in myeloid immortalization assays. (D) Expression levels of Hoxa9, Meis1, Evi1, and MLL-ENL by QRT-PCR in the immortalization assays. Tg LT-HSCs (CD34[−]) and ST-HSCs (CD34[+]) retrovirally transduced with CreER were harvested at the end of each round, and the first round, of plating, respectively. (E-F) Colony replating assays of subpopulations in the immortalized cells constituting colonies of CreER-transduced Tg LT-HSCs after the third round of plating, sorted on the basis of the immunophenotype (E). (G) Immunophenotype of the cells derived from the c-Kit+Sca-1+Gr-1– subpopulation of the immortalized Tg cells, harvested at the first replating. The bar graphs show the mean ± SD of 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/7/10.1182_blood-2012-09-456665/4/m_1271f2.jpeg?Expires=1768288610&Signature=RmSRp9eK4k2ZyzCrRbDj2HwMpaVQEFXFTRZGHTV15u1wFwtBhGQStw3h8Zhtfyty~tGn6O8f4pcgIV-boENIw4qWALsDmoLSO9ouTzTRvIUAfIySKGOWbxQYlfdGW4CPyJ4VW-VNH-glnQLg0tiP6EE3XJ0Z4m0s1bFaXKpQyn66--4aCHpXiLhsohy~yZ9b3aPKW9p8Q-nQg9k962JnKgbX0ikpSIJ-SbA5PXg1GI4eBIFVKcCI~hRZyapc04k8HBO2vDS7mDYJQqA-QdYDojFrPh0LMVCAGHZMj4YoZG8vc7AEz95i7G-gT9Yg-kPoi6YjslUl02ZWINjpkiCaBg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. Myeloid immortalization of HSCs by MLL fusion genes associated with high Plzf expression. (A) Expression level of Plzf by QRT-PCR in myeloid immortalization assays. Tg LT-HSCs (CD34[−]), and ST-HSCs (CD34[+]) and MPs, retrovirally transduced with mock or CreER were harvested at the end of each round, and the first round, of plating, respectively. (B) Intracellular FACS analyses of Plzf expression in the immortalized cells constituting colonies of the CreER-transduced Tg LT-HSCs after the third round of plating. (C) Expression levels of Plzf by QRT-PCR in WT whole KSL or MP cells retrovirally transduced with mock, MLL-ENL, or MLL-SEPT6 at the first replating in myeloid immortalization assays. (D) Structure of Plzf (BTB/POZ domain and zinc fingers [Zn]) and a mutant (PlzfΔBTB) lacking the BTB/POZ domain, and expression of Plzf and the mutant by Western blot (lower left panels) and RT-PCR (lower right panels) analyses. Lysates extracted from Plat E cells transfected with mock (pMYs-IP), pMYs-Plzf-IP, or pMYs-PlzfΔBTB-IP were blotted with the anti-Plzf (αPlzf) antibody, followed by reprobe with the anti–α-tubulin antibody (αTub) as an internal control. The WT whole KSL and MP cells retrovirally transduced with mock, Plzf, or PlzfΔBTB were harvested at the first replating in myeloid immortalization assays and were subjected to RT-PCR. B2m was used as an internal standard. NTC, no template control. (E) Myeloid immortalization assays of WT LT-HSCs (CD34[−]), ST-HSCs (CD34[+]), whole KSL cells, and MPs retrovirally transduced with mock, Plzf, or PlzfΔBTB by replating 104 cells. (F-H) Typical morphology of the colonies (F) in the Plzf-transduced KSL cells at the end of the third round of plating, and typical morphology (G) and immunophenotype (H) of the cells constituting these colonies. Colonies, and cells stained with Wright-Giemsa, were viewed with an Olympus CKX41 microscope using a 4×/0.13 objective lens and an Olympus BX41 microscope using a 20×/0.5 objective lens, respectively. Magnification and bars in panel F indicate 40× and 200 μm; magnification and bars in (G) indicate 200× and 20 μm. (I) Expression levels of Hoxa9, Meis1, and Evi1 in the immortalization assays, compared with the CreER-transduced Tg LT-HSCs (CD34[−]) by QRT-PCR. WT whole KSL and MP cells were retrovirally transduced with mock or Plzf and harvested at the end of the first and third rounds of plating. The CreER-transduced Tg LT-HSCs were harvested at the end of the third round of plating. The bar graphs show the mean ± SD of 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/7/10.1182_blood-2012-09-456665/4/m_1271f3.jpeg?Expires=1768288610&Signature=4Vby46mJ~WtVhF8nT0gZQhlw7V5N6A2dWGl6R0Gopkq9okPE1yy7wLTnlETLRy-z4rr2~HKeXEOKXW5f3nKRz540J5wwxInX60FMk94R7wxzgFTX6jy7l34xo14LGaaQcdJyvhJQQIxUbz9lBdzmWLG9jcAXONWdnydptiFk--A0LussWSkweHZYbF6Birh8A7Fae-z52myjGQhWDgdL3HadfnuDFxCExN~s7T8xl7KGvvtG7pEHiuJn4T23IfsHu6F~47nZ-T9pt5ujqsKslfWTSww8Ronf3VST0Hgn9ZlsH~7b6xAwJi8rDnSFs4ZKgS4zp5mvL6N~QtNbcTR6XA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 5. Rescue of MLL-fusion–immortalized KSL cells from Plzf depletion by transduction with shRNA-resistant Plzf, but not with a resistant Plzf lacking the BTB/POZ domain. (A) Expression levels of Plzf by QRT-PCR (upper) and relative CFU (lower) of the cells sorted from shRNA-transduced immortalized cells in colony forming assays. WT whole KSL and MP cells immortalized by retroviral transduction of MLL-ENL, MLL-SEPT6, or E2A-HLF were harvested at the end of the first plating. (B) Structures of Plzf and a mutant lacking the BTB/POZ domain, modified with introduction of silent mutations (*) resistant to shRNA (shP01) against Plzf. Arrows indicate primers to detect Plzf transcripts in the coding region. (C) Expression levels of the mutated Plzf by QRT-PCR in the MLL-ENL–immortalized WT KSL (ME) cells retrovirally transduced with Plzf* (ME+Plzf*) or Plzf*/ΔBTB (ME+Plzf*/ΔBTB). (D-E) Expression levels of Plzf by QRT-PCR (D) and relative CFU (E) of the cells sorted from shRNA-transduced ME+Plzf* or ME+Plzf*/ΔBTB cells in colony replating assays. QRT-PCR analyses were performed using primers in the coding region (A [upper], C,D [upper]) or the 3′ untranslated region (D [lower]). The bar graphs show the mean ± SD of 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/7/10.1182_blood-2012-09-456665/4/m_1271f5.jpeg?Expires=1768288610&Signature=3VVjlRqddD9gp2JhKBw7igFBHXapSXKaTGDHv0PqzW6W0oAXwe-fZZSWWTZ-hYXqPyEoTSQPVAe7PGI1cXJRGmUQa3PLfaLOENV70kKChmkVaXFVvhnjTqL6Neu9c4b-P6xyo5UgJXAXAxvXSn76GFJuCt~gvxYwHzNm9U~XBa34ccAcizD0OyRwDbsOjSCGhmyKn0Aj15FHK6epm~MfsiH8LzxASyd1mCA2FFPnNNavqBP3Hb4T-mQP2lNUQnwV4NNJfwlxcF7xUoCClgMwz9k7un5Z1h2mAwpJmdvzl7EMdVtUV8l27V1vrGyNLNqblqXRXQ1LKNaGu13oJ49Xgw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 1. Leukemic transformation of Tg LT-HSCs conditionally expressing MLL-ENL. (A) Construction of the plasmid for conditional expression of MLL-ENL. CAG, hybrid CMV enhancer/chicken β-actin promoter; FLAG, tag sequence fused to the C terminus; SV40, simian virus 40. Arrowheads indicate loxP sites; filled diamond indicates triple stop codon sequence; horizontal arrows indicate primers to detect recombination between the loxP sites in (D); vertical arrow indicates the EcoR I site. (B) Expression of GFP in peripheral white blood cells from a conditional MLL-ENL Tg mouse (black line) compared with a WT littermate (gray shadow) on FACS. (C) Experimental strategy for myeloid immortalization assays and BMT. Sorted BM cells were retrovirally transduced with transgenes (dotted diagonal lines). (D) Recombination detected by genomic PCR in myeloid immortalization assays of WT and Tg LT-HSCs retrovirally transduced with mock (harvested at the end of the first plating) or CreER (Tg only; harvested at the end of each round of plating). G, germline configuration; R, recombination between loxP sites. (E) Expression levels of Hoxa9, Meis1, Evi1, and MLL-ENL by QRT-PCR in myeloid immortalization assays. Tg LT-HSCs (CD34[−]), Tg ST-HSCs (CD34[+]), Tg MPs, WT whole KSL cells, and WT MPs were retrovirally transduced with mock, CreER, or MLL-ENL and harvested at the first replating. (F) Myeloid immortalization assays of cells with retroviral transduction, as described in (E), by replating 104 cells. (G-H) Typical morphology of the colonies of the Tg LT-HSCs retrovirally transduced with CreER at the end of the third round (G), and the immortalized cells constituting the colonies (H). Colonies, and cells stained with Wright-Giemsa, were viewed with an Olympus CKX41 microscope using a 4×/0.13 objective lens and an Olympus BX41 microscope using a 20×/0.5 objective lens, respectively. Images were acquired with Olympus DP21 and Olympus DP21 software. Magnification and bars in (G) indicate 40× and 200 μm; magnification and bars in (H) indicate 200× and 20 μm. (I) Immunophenotype of the immortalized Tg LT-HSCs (CD34–KSL) and the WT whole KSL cells immortalized by retroviral transduction of MLL-ENL. (J) Survival curves of mice transplanted with Tg LT-HSCs (CD34–; n = 4), ST-HSCs (CD34+; n = 4), whole KSL cells (n = 5 for CreER; n = 3 for mock), and MPs (n = 4), retrovirally transduced with CreER(-IRES-EGFP) or mock. The bar graphs indicate the mean ± SD of 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/7/10.1182_blood-2012-09-456665/4/m_1271f1.jpeg?Expires=1768298970&Signature=wln89zLq9xSnwxVGBP12Ap0alVelxXYgodVb43tsdzqG~Hg4icPGeU2kiEjUgDBGcqQLtU0HIEhembjtxWbv6AWs~bnvrOnkLW2AzHXWIiWa7Gxb4vjEciVYJdQG5arqgL9Gi1idecDHXYkMV9pajsSH3uvqai0-TDncOrjK9PZuR~CkJqI3BX48mgcwee7knfOrhmHZ1PFTfaTqfpfYqxWuZTrYFLtlo9oryQ-4H9hyoKbmY5Y0MyXS3ZGl2PjsJzMxRazI1rWM1bHgDUFEMBQGmfE6izymwCz4qT8SUDpT0Aew3UXisGdjDEOY1timb-MVGNayKlodFlqFKkRSiQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. Characterization of Tg LT-HSCs conditionally expressing MLL-ENL. (A) Schematic representation of human MLL-ENL (MLL: thick; ENL: thin) transcript and the corresponding region of the endogenous transcript of mouse Mll. Mll and the total MLL-ENL/Mll transcripts were analyzed by QRT-PCR using common primers (horizontal arrows) with either Mll- or MLL/Mll-TaqMan probes. (B-C) Expression level of Mll (B) and relative ratio of total MLL(-ENL)/Mll to Mll transcripts (C), quantified by QRT-PCR. Tg LT-HSCs (CD34[−]), ST-HSCs (CD34[+]), and MPs retrovirally transduced with mock or CreER were harvested at the first replating in myeloid immortalization assays. (D) Expression levels of Hoxa9, Meis1, Evi1, and MLL-ENL by QRT-PCR in the immortalization assays. Tg LT-HSCs (CD34[−]) and ST-HSCs (CD34[+]) retrovirally transduced with CreER were harvested at the end of each round, and the first round, of plating, respectively. (E-F) Colony replating assays of subpopulations in the immortalized cells constituting colonies of CreER-transduced Tg LT-HSCs after the third round of plating, sorted on the basis of the immunophenotype (E). (G) Immunophenotype of the cells derived from the c-Kit+Sca-1+Gr-1– subpopulation of the immortalized Tg cells, harvested at the first replating. The bar graphs show the mean ± SD of 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/7/10.1182_blood-2012-09-456665/4/m_1271f2.jpeg?Expires=1768298970&Signature=dCgY~~3k2SyCKheoIrSYNOYKgC2djLOnyP24SJhsuWc8hj4J8euj6GBjiDGxR1KWJqmL9PkL9WlE80O0NK0I01u68Vt1Vch7Ku4DfSL7Hi9bwcWOdeJ9WmrtCEN6AZ6s-Su0jWtb5OCuPrVTt3svM-WyEndrN5qjWdSAD~9nNf3RB24zSbF8~87ytRHnDw5-amkAp4KmBAfB2FRdb6hURjSdmV-4-mEtp2d-JUiiZTZKPY2AUPV7NGKgmYxoURqMKs02x2knz41Hf3v4dqDioyOLYu-ujezVUIVjOjNZPdxOvoj3ruhle0gu8ei7VXfhCpOG5WCkaYy8xFGtpsQTbg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. Myeloid immortalization of HSCs by MLL fusion genes associated with high Plzf expression. (A) Expression level of Plzf by QRT-PCR in myeloid immortalization assays. Tg LT-HSCs (CD34[−]), and ST-HSCs (CD34[+]) and MPs, retrovirally transduced with mock or CreER were harvested at the end of each round, and the first round, of plating, respectively. (B) Intracellular FACS analyses of Plzf expression in the immortalized cells constituting colonies of the CreER-transduced Tg LT-HSCs after the third round of plating. (C) Expression levels of Plzf by QRT-PCR in WT whole KSL or MP cells retrovirally transduced with mock, MLL-ENL, or MLL-SEPT6 at the first replating in myeloid immortalization assays. (D) Structure of Plzf (BTB/POZ domain and zinc fingers [Zn]) and a mutant (PlzfΔBTB) lacking the BTB/POZ domain, and expression of Plzf and the mutant by Western blot (lower left panels) and RT-PCR (lower right panels) analyses. Lysates extracted from Plat E cells transfected with mock (pMYs-IP), pMYs-Plzf-IP, or pMYs-PlzfΔBTB-IP were blotted with the anti-Plzf (αPlzf) antibody, followed by reprobe with the anti–α-tubulin antibody (αTub) as an internal control. The WT whole KSL and MP cells retrovirally transduced with mock, Plzf, or PlzfΔBTB were harvested at the first replating in myeloid immortalization assays and were subjected to RT-PCR. B2m was used as an internal standard. NTC, no template control. (E) Myeloid immortalization assays of WT LT-HSCs (CD34[−]), ST-HSCs (CD34[+]), whole KSL cells, and MPs retrovirally transduced with mock, Plzf, or PlzfΔBTB by replating 104 cells. (F-H) Typical morphology of the colonies (F) in the Plzf-transduced KSL cells at the end of the third round of plating, and typical morphology (G) and immunophenotype (H) of the cells constituting these colonies. Colonies, and cells stained with Wright-Giemsa, were viewed with an Olympus CKX41 microscope using a 4×/0.13 objective lens and an Olympus BX41 microscope using a 20×/0.5 objective lens, respectively. Magnification and bars in panel F indicate 40× and 200 μm; magnification and bars in (G) indicate 200× and 20 μm. (I) Expression levels of Hoxa9, Meis1, and Evi1 in the immortalization assays, compared with the CreER-transduced Tg LT-HSCs (CD34[−]) by QRT-PCR. WT whole KSL and MP cells were retrovirally transduced with mock or Plzf and harvested at the end of the first and third rounds of plating. The CreER-transduced Tg LT-HSCs were harvested at the end of the third round of plating. The bar graphs show the mean ± SD of 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/7/10.1182_blood-2012-09-456665/4/m_1271f3.jpeg?Expires=1768298970&Signature=Y4C5eq8GgoKXRATXQlCo9Ah-XVJPaoE7gFq0nCGA5tSouKSaw-WTvBpVIW4Dxzlgt7Pnis-k~Mo~Sgfi2EKpyrfd78yhXGMTTF4LI2f~9cpwAcjHyyxqBRfuKNjwCy7a-AqhEv006F8aOhmeEVtjxUKniCslCIKhHZNj8H6ZKRGZ9ftn497lP8iJAEPVVZwSi1L9uVP6ams7xj9lQiG6MtBqNNVp-Itathsd6pI3rDB-QTPODLJaRV90hrFsowbDpcRXMS4n-oT44LDO6RjzmV6Yw~yqmvICSE2cZz9pVGZW0IaMTCxy05KF3CeZri3UBAeZxNj35-yb31RXo7ipCA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. Rescue of MLL-fusion–immortalized KSL cells from Plzf depletion by transduction with shRNA-resistant Plzf, but not with a resistant Plzf lacking the BTB/POZ domain. (A) Expression levels of Plzf by QRT-PCR (upper) and relative CFU (lower) of the cells sorted from shRNA-transduced immortalized cells in colony forming assays. WT whole KSL and MP cells immortalized by retroviral transduction of MLL-ENL, MLL-SEPT6, or E2A-HLF were harvested at the end of the first plating. (B) Structures of Plzf and a mutant lacking the BTB/POZ domain, modified with introduction of silent mutations (*) resistant to shRNA (shP01) against Plzf. Arrows indicate primers to detect Plzf transcripts in the coding region. (C) Expression levels of the mutated Plzf by QRT-PCR in the MLL-ENL–immortalized WT KSL (ME) cells retrovirally transduced with Plzf* (ME+Plzf*) or Plzf*/ΔBTB (ME+Plzf*/ΔBTB). (D-E) Expression levels of Plzf by QRT-PCR (D) and relative CFU (E) of the cells sorted from shRNA-transduced ME+Plzf* or ME+Plzf*/ΔBTB cells in colony replating assays. QRT-PCR analyses were performed using primers in the coding region (A [upper], C,D [upper]) or the 3′ untranslated region (D [lower]). The bar graphs show the mean ± SD of 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/7/10.1182_blood-2012-09-456665/4/m_1271f5.jpeg?Expires=1768298970&Signature=O8nc4gtxM73lz5NIH7GW5To8CRb5NXhjLNhasE3jjlcRxUwHwMTxkYUwQyWkv5tIYiMhSdzd9JXmBCG0cTXQACp~OXjMR4ghhltd0ful~mWDtwXDVHRBSfsqdTL3iiDBTuq7oaseg39kqfU0lBL7jZKqYBQbpcLAlpHHzNkSFKC3hiPCs6DVYjkuwwbWd5323RzcUKTRewVpic9dAA3rB3epcu0TEH5CBaN7n~BGA8OeCvXhEyr346ZobV3AzB42wkzAFVthe4ht1YrCSXQMy1BeuBani~CJssaQip~0yYSEiFiYbvD7vfhbMjrxqtukQ7tR~gR~W6w4q5CCRyJf2w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)