In this issue of Blood, Mleczko-Sanecka et al used a novel approach based on genome-wide RNA interference knockdown technology to identify the genes involved in the regulation of hepcidin. It showed that hepcidin suppression is linked to the control of mitogen stimulation and nutrient status via components of well-characterized signaling pathways. This indicates novel links between the control of systemic iron homeostasis and critical liver processes such as regeneration, response to injury, carcinogenesis, and nutrient metabolism.1
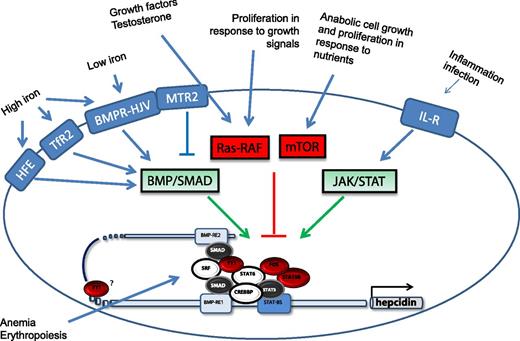
Scheme of the stimuli that modify liver hepcidin expression and of the molecules and signal transduction pathways involved. The newly identified molecules (shown in red) are described in Mleczko-Sanecka et al.1 Adapted from Figure 6 in the article by Mleczko-Sanecka et al that begins on page 1574.
Scheme of the stimuli that modify liver hepcidin expression and of the molecules and signal transduction pathways involved. The newly identified molecules (shown in red) are described in Mleczko-Sanecka et al.1 Adapted from Figure 6 in the article by Mleczko-Sanecka et al that begins on page 1574.
Hepcidin is a peptide hormone mainly synthesized in the liver that controls systemic iron availability and adjusts it to tissue requirements.2 It binds its receptor with high affinity, inducing its fast internalization and degradation. Since the receptor is the cellular iron exporter ferroportin, the consequence of the binding is to block iron export to transferrin and its transport to the tissues.2 Hepcidin expression is controlled mainly at a transcriptional level by various stimuli, including iron status, hypoxia, erythropoietic activity, and inflammation. Most iron-related disorders are attributed to a deregulation of liver hepcidin expression. These disorders include the various forms of hereditary hemochromatosis, which is associated with low hepcidin expression or functionality,3 and genetic iron-refractory iron deficiency anemia, which is associated with high hepcidin levels and iron restriction.4
Most of our understanding of hepcidin regulation derives from clinical studies and animal models. For example, the centrality of the bone morphogenetic protein/sons of mothers against decapentaplegic (BMP/SMAD) pathway originated from the observation that liver conditional ablation of SMAD4 fully suppressed hepcidin expression and caused severe iron overload.5 Interestingly, the article by Mleczko-Sanecka et al1 supports and reinforces this centrality. The crucial involvement of hemojuvelin (HJV) and of the protease that controls its functionality (TMPRSS6 or matriptase-2) was indicated by genetic studies. Similar approaches identified several other genes that participate in hepcidin control, including transferrin receptor 2 (TfR2), the inhibitory SMAD7, hypoxia inducible factor, erythropoietin, soluble factors of the transforming growth factor beta family, growth factors, and testosterone. Among them is HFE, the major cause of hereditary hemochromatosis. Although frequently studied, it has not been fully clarified how HFE affects the BMP-HJV-SMAD pathway and regulates hepcidin expression and which are the genetic and/or environmental modifiers that modify the penetrance of the C282Y mutation of HFE. This genome-wide, unbiased methodology used by the Muckenthaler-Hentze team goes some way to identify such genetic factors (see figure).
The approach used is original and is carried out in a rigorous way. Mleczko-Sanecka et al used a library of small interfering RNA pools targeting almost 20 000 human genes. It was applied in a high-throughput system to modulate the activity of the hepcidin promoter-luciferase reporter system. The response range of interest was defined in such a way that it identified 1651 putative activators and 508 putative inhibitors, which included several of the known activators and inhibitors. This long list of putative hepcidin regulators is enriched for genes involved in signal transduction, transcriptional regulation, and defense and/or inflammatory responses and is a precious source for future analyses. Bioinformatics and literature mining were used for the next difficult step to restrict the analysis to a limited number of genes. Genes involved in signal transduction and transcriptional regulation were chosen, and 15 (25%) of them were validated in stringent secondary assays. Further analysis showed that most of the genes needed functional BMP/SMAD responsive elements to regulate the hepcidin promoter, while none were affected by the inactivation of the inflammatory interleukin-6 (IL-6) responsive element. Next, they found that the downregulation of the genes caused pronounced alteration of proteins controlling the rat sarcoma-mitogen activated protein kinase (Ras-MAPK) and rho signaling (Ras homolog signaling), which are interconnected with β-catenin and mammalian target of rapamycin (mTOR) signaling. The involvement of these pathways was verified by specific drug inhibitors, such as sorafenib, wortmannin, rapamycin, and metformin. In various hepatic cells, hepcidin messenger RNA was induced by these drugs and, conversely, it was experimentally suppressed by the stimulation of the pathways.
The study has a number of important implications. First, it confirmed that the BMP-HJV-SMAD signaling pathway is the core axis of hepcidin control and supports it to be the major target for pharmacologic control of hepcidin expression.6 Second, it shows that the signaling can be modified by cross-talking with other pathways, including well-characterized ones such as the nutrient-sensing mTOR and proliferative Ras/RAF pathways. This may help explain the known effects of growth factors, cytokines, and liver stress conditions on hepcidin expression. Probably more important is that some of these pathways are the targets of widely used drugs, and their effects may be tested in vivo in animal models with abnormal hepcidin expression. For example, sorafenib is in clinical use for treatment of hepatocellular carcinoma, and its capacity to upregulate hepcidin may cause local iron restriction and may contribute to its antitumor activity.7 A recent retrospective clinical study showed that the use of the mTOR inhibitor rapamycin is associated with mild microcytic anemia, likely attributable to iron restriction.8 It remains to be demonstrated that this is linked to hepcidin excess, as predicted by Mleczko-Sanecka et al.1
In conclusion, this work leave us with an elegant and rigorous approach to finding genes involved in hepcidin regulatory pathways. It is expected to stimulate future work to characterize the role of the newly identified signaling pathways in the pathogenicity of iron-related diseases.
Conflict-of-interest disclosure: The author declares no competing financial interests.