Abstract
Most proteins in nature are chemically modified after they are made to control how, when, and where they function. The 3 core features of proteins are posttranslationally modified: amino acid side chains can be modified, peptide bonds can be cleaved or isomerized, and disulfide bonds can be cleaved. Cleavage of peptide bonds is a major mechanism of protein control in the circulation, as exemplified by activation of the blood coagulation and complement zymogens. Cleavage of disulfide bonds is emerging as another important mechanism of protein control in the circulation. Recent advances in our understanding of control of soluble blood proteins and blood cell receptors by functional disulfide bonds is discussed as is how these bonds are being identified and studied.
Introduction
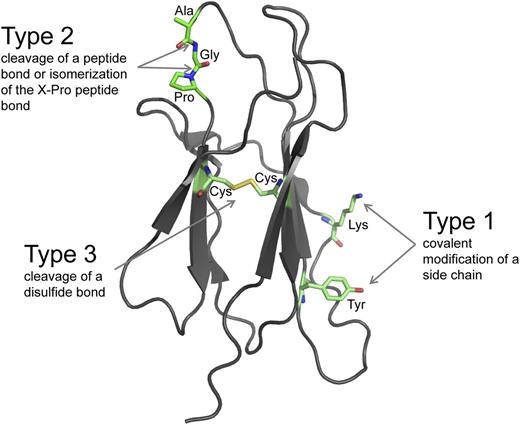
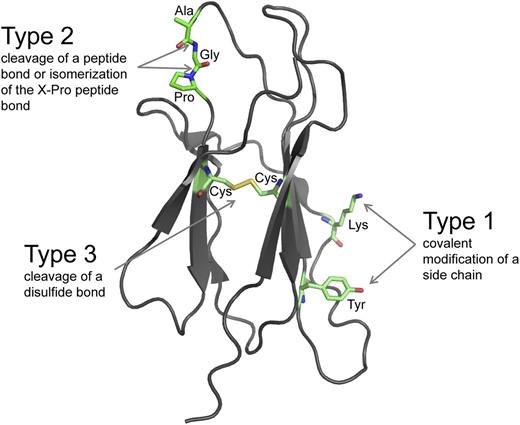
There are 3 fundamental types of posttranslational modifications of proteins that appear to operate in all life forms.1-3 Amino acid side chains can be covalently modified (type 1), peptide bonds can be hydrolytically cleaved or isomerized (type 2), and disulfide bonds can be reductively cleaved (type 3) (Figure 1). These chemical changes are involved in all aspects of protein function and vastly increase the sophistication of the proteome.
The 3 fundamental types of posttranslational modifications. The ribbon structure of a model protein is shown. The amino acid side chains and the peptide and disulfide bonds that bind the polypeptide backbone can be posttranslationally modified. Type 1 modifications are covalent additions of a molecule to an amino acid side chain. The side chains of 15 of the 20 common amino acids of proteins can be covalently modified in reactions that usually involve an enzyme and a cosubstrate.1 The lysine and tyrosine side chains are shown. The most frequent Type 1 modification in humans is phosphorylation of tyrosine. Type 2 modifications are hydrolytic cleavage or isomerization of certain peptide bonds. Hydrolytic cleavage is catalyzed by proteases, which are tightly regulated in space and time because the cleavage is irreversible. Isomerization of the peptide bond on the C-terminal side of proline residues is catalyzed by peptidyl prolyl cis-trans isomerases. Type 3 modifications are reductive cleavage of certain disulfide bonds, known as allosteric disulfides. Allosteric disulfide bonds control the function of the mature protein in which they reside by mediating a change when they are cleaved by oxidoreductases or by thiol-disulfide exchange.
The 3 fundamental types of posttranslational modifications. The ribbon structure of a model protein is shown. The amino acid side chains and the peptide and disulfide bonds that bind the polypeptide backbone can be posttranslationally modified. Type 1 modifications are covalent additions of a molecule to an amino acid side chain. The side chains of 15 of the 20 common amino acids of proteins can be covalently modified in reactions that usually involve an enzyme and a cosubstrate.1 The lysine and tyrosine side chains are shown. The most frequent Type 1 modification in humans is phosphorylation of tyrosine. Type 2 modifications are hydrolytic cleavage or isomerization of certain peptide bonds. Hydrolytic cleavage is catalyzed by proteases, which are tightly regulated in space and time because the cleavage is irreversible. Isomerization of the peptide bond on the C-terminal side of proline residues is catalyzed by peptidyl prolyl cis-trans isomerases. Type 3 modifications are reductive cleavage of certain disulfide bonds, known as allosteric disulfides. Allosteric disulfide bonds control the function of the mature protein in which they reside by mediating a change when they are cleaved by oxidoreductases or by thiol-disulfide exchange.
The type 1 modifications of side chains nearly always require an enzyme and a cosubstrate, so these events are usually restricted to specific intracellular environments where all 3 components are available.1 Type 2 modifications of peptide bonds and type 3 modifications of disulfide bonds are suited to the circulation. Cleavage of peptide or disulfide bonds usually does not require a cofactor, so the factors that mediate these events need only to find their substrate to function.
Disulfide bonds in blood proteins
Protein disulfide bonds are the links between the sulfur atoms of 2 cysteine amino acids (the cystine residue) that form as proteins mature in the cell. These bonds have accrued during the evolution of eukaryotic proteins and, once acquired, have almost always been retained.4
The tertiary structures or partial tertiary structures of 4104 human proteins are currently known or inferred from a homologous protein/domain; these contain 16 538 disulfide bonds (UniProt annotation). About half the disulfide bonds (7264) are in membrane proteins (1987) and most of the rest (8424) are in proteins containing a secretion signal sequence (1204). Interestingly, 587 proteins that reside in the cytoplasm or nucleoplasm contain 935 disulfide bonds, which are environments traditionally thought not to be conducive to disulfide bond formation. The mechanism of formation of disulfide bonds in cytoplasm and nucleoplasm proteins is largely unknown, although protein disulphide isomerase (PDI) has recently been found to interact with the actin cytoskeleton.5 These numbers are representative of the protein disulfide bonds in and on leukocytes.
Mass spectrometry analysis of human plasma has identified 1929 different proteins.6 The tertiary structures of 817 of these proteins are currently known or inferred from a homologous structure/domain; these contain 4594 disulfide bonds. Assuming this average protein to disulfide bond ratio of 1:5 holds for the remaining proteins with unknown structure, the 2000 or so plasma proteins will contain about 10 000 disulfide bonds.
Most of these disulfide bonds, as with most of the peptide bonds, perform a structural role. They stabilize the mature protein structure and remain unchanged for the life of the protein. However, some of the disulfide bonds—the allosteric disulfides—control the function of the mature protein in which they reside when they are cleaved.
Functional disulfides
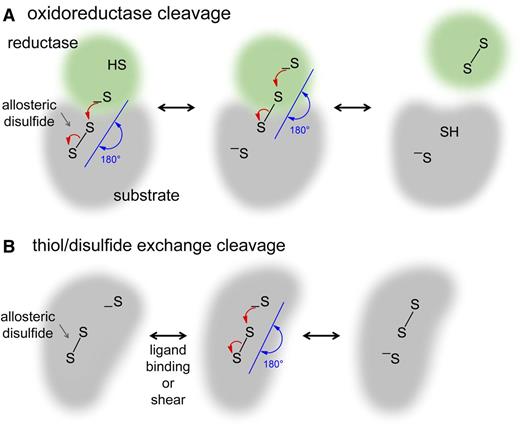
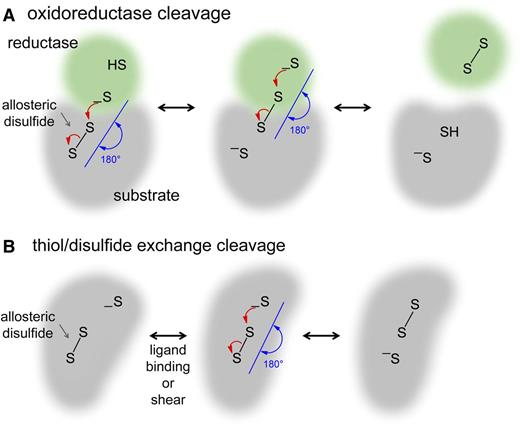
Allosteric control is defined as a change in 1 site—the allosteric site—that influences another site by exploiting the protein's flexibility.7 Thus, cleavage of an allosteric disulfide bond results in a functional change at another site in the protein. Changes in ligand binding, substrate hydrolysis, proteolysis, or oligomer formation have been identified in blood proteins.2 The allosteric disulfide bonds are reduced by the catalytic disulfides of oxidoreductases (Figure 2A) or by thiol-disulfide exchange (Figure 2B).2,8
Mechanisms of cleavage of allosteric disulfide bonds. Disulfide bond reduction occurs via a second-order nucleophilic substitution (SN2)-type reaction mechanism in which the 3 sulfur atoms involved must form an ∼180° angle. (A) For oxidoreductase cleavage, the active site sulfur ion nucleophile of the oxidoreductase (green) attacks 1 of the sulfur atoms of the allosteric disulfide bond (gray). The mixed disulfide that forms then spontaneously decomposes, releasing oxidized oxidoreductase and the substrate protein containing a reduced allosteric disulfide. (B) For thiol-disulfide exchange, the protein contains a sulfur ion nucleophile that is unreactive until a conformational change brings the sulfur ion in line with the allosteric bond, where it attacks 1 of the sulfur atoms of the disulfide, cleaving the bond. The conformational change can be mediated by ligand binding or by mechanical shear of the protein. Intramolecular cleavage is shown, but this can also occur intermolecularly.
Mechanisms of cleavage of allosteric disulfide bonds. Disulfide bond reduction occurs via a second-order nucleophilic substitution (SN2)-type reaction mechanism in which the 3 sulfur atoms involved must form an ∼180° angle. (A) For oxidoreductase cleavage, the active site sulfur ion nucleophile of the oxidoreductase (green) attacks 1 of the sulfur atoms of the allosteric disulfide bond (gray). The mixed disulfide that forms then spontaneously decomposes, releasing oxidized oxidoreductase and the substrate protein containing a reduced allosteric disulfide. (B) For thiol-disulfide exchange, the protein contains a sulfur ion nucleophile that is unreactive until a conformational change brings the sulfur ion in line with the allosteric bond, where it attacks 1 of the sulfur atoms of the disulfide, cleaving the bond. The conformational change can be mediated by ligand binding or by mechanical shear of the protein. Intramolecular cleavage is shown, but this can also occur intermolecularly.
Oxidoreductase cleavage
Oxidoreductase active sites contain a reactive dithiol/disulfide in a CXXC motif that can reduce or oxidize a substrate disulfide bond (Figure 2A). Seven members of the PDI family,9 2 members of the thioredoxin family, and glutaredoxin-110 have been identified in human plasma6 (Table 1). Three other members of the PDI family, transmembrane TMX3, ERp44, and ERp29, and the flavoenzyme, Ero1α, have been detected on the platelet surface.11,12 ERp44 and ERp29, however, do not contain a CXXC active site motif, so they may not participate in thiol-disulfide exchange reactions.9 The roles in the circulation of the PDI family members—PDI and ERp57—are best understood to date. There is also emerging evidence for a role for ERp5 in platelet function and thioredoxin in the inflammatory response.
PDI is secreted by platelets,13 endothelial cells,14,15 and neutrophils,16 and mediates protein dithiol-disulfide exchange events in the extracellular space.17,18 This oxidoreductase binds to β3 integrins in the thrombus19 and αvβ3 integrin on endothelial cells.20 PDI inhibitors reduce platelet thrombus size and fibrin formation in both micro- and macrovascular murine models.14,21-23 Platelet PDI is involved in αIIbβ3 integrin–mediated platelet accumulation,24 whereas endothelial PDI is required for fibrin formation.14 Neutrophil PDI modulates ligand binding to αMβ2 integrin and neutrophil recruitment during venous inflammation.25
The PDI family member ERp57 translocates to the platelet surface after activation, binds to β3 integrin, and is involved in platelet activation and aggregation in vitro and incorporation of platelets into a growing thrombus in murine models.26,27 PDI and ERp57 appear to have distinct roles in β3 integrin function and platelet aggregation as addition of function blocking anti-ERp57 antibodies to PDI-deficient platelets further diminishes integrin αIIbβ3 activation and platelet aggregation.24 Another PDI family member, ERp5, is also recruited to the platelet surface after activation in vitro, and function-blocking anti-ERp5 antibodies inhibit platelet aggregation, fibrinogen binding, and P-selectin exposure.28 A role for ERp5 in platelet function in vivo has not been reported to date.
Thioredoxin has been implicated in the inflammatory response in the circulation. Expression of this reductant is upregulated, and the protein is secreted by immune cells during inflammation, leading to a high local concentration and elevated blood levels.29 Serum levels of thioredoxin are increased in patients with asthma, rheumatoid arthritis, and heart failure30,31 and positively correlate with disease activity in rheumatoid arthritis.32,33
PDI, ERp57, possibly ERp5, and thioredoxin are involved, therefore, in thrombosis and/or inflammation in mammals. The target disulfide bonds of these oxidoreductases are being defined. PDI is involved in reduction or formation of disulfide bonds in some platelet and leukocyte integrins and possibly leukocyte tissue factor (TF) (see the following section). Thioredoxin reduces the allosteric disulfides in β2-glycoprotein I34,35 and mast cell β tryptase36 in vitro; the role of this reductase in vivo is being studied (see the following section).
The general question of which oxidoreductase cleaves which allosteric disulfide will be determined by some of the same factors that govern which protease cleaves which peptide bond. Some proteases cleave many peptide bonds (such as thrombin), whereas others cleave only 1 or a very restricted number (such as factor VIIa), and the same scenario will likely apply to oxidoreductases and allosteric disulfide bonds. Oxidoreductase substrate selectivity will largely be driven by steric factors (accessibility of the disulfide bond in the substrate protein and conformation of the active site pocket of the enzyme) and by environmental context (being in the right place at the right time). The highly directional nature of disulfide bond cleavage will further restrict the substrate selection of oxidoreductases. Reduction of a disulfide bond proceeds via a second-order nucleophilic substitution (SN2)-type reaction mechanism in which the 3 sulfur atoms involved (the sulfur ion nucleophile and the 2 sulfur atoms of the disulfide bond) must form an ∼180° angle37,38 (Figure 2). For instance, stretching, twisting, or pulling of proteins makes their disulfide bonds easier or harder to cleave by changing the alignment of the 3 sulfur atoms.39,40 Another factor that influences the substrate selectivity of oxidoreductases is the redox potential of the oxidoreductase catalytic disulfide and the allosteric disulfide. Catalytic disulfides will only cleave allosteric disulfides with a bigger (less negative) redox potential.
An open question is whether the oxidoreductases act catalytically in the circulation to reduce several substrate disulfides or reduce only 1 disulfide bond. Intracellular oxidoreductases act as catalysts because factors regenerate the reduced enzyme. The thioredoxin active site disulfide, for example, is reduced by thioredoxin reductase and reduced NAD phosphate (NADPH) in the cytoplasm.10 NADPH provides the hydrogens and electrons used to reduce the oxidized thioredoxin. Thioredoxin reductase is found in plasma,6 but NADPH is not. It is possible, and seems likely, that a mechanism or mechanisms exist to cycle some oxidoreductases in blood. For instance, Ero1α has been implicated in oxidation of PDI on the platelet surface.12 On the other hand, restricting oxidoreductases to cleavage of a single substrate disulfide bond would be an efficient mechanism of regulation of their activity. Proteases, for instance, only require a source of water to cleave peptide bonds, so protease inhibitors have coevolved with proteases to control their activity.1 Controlling access to a source of hydrogens and electrons is a simple means of restricting oxidoreductase activity. Endogenous inhibitors of the oxidoreductases in the extracellular space have not been identified to date, which supports the idea that oxidoreductases may function as single-turnover enzymes.
Thiol-disulfide exchange cleavage
Cleavage of allosteric disulfides by thiol-disulfide exchange does not require additional hydrogens and electrons (Figure 2B), so this mechanism of cleavage is particularly suited to the circulation. All that is required is a conformational change in the substrate protein, which could be triggered by ligand binding and/or the mechanical shear forces of flowing blood. The substrate protein contains a sulfur ion that is unreactive until a change in structure brings it in line with the allosteric bond, where it attacks and cleaves the disulfide bond. Allosteric disulfide bonds in plasma plasminogen41 and von Willebrand factor (VWF)42 are cleaved by thiol-disulfide exchange (see the following section).
An important difference between peptide and disulfide bond cleavage is that cleavage of disulfide bonds is potentially reversible; this can provide for a fine level of protein regulation not possible with peptide bond cleavage. Some allosteric disulfides exist in equilibrium between reduced and oxidized isoforms in the population of protein molecules, and perturbation of the equilibrium by oxidoreductase or ligand binding, for instance, leads to a biological change. Cleavage of other allosteric disulfides, though, is irreversible as the downstream effects of the cleavage prevent the disulfide bond from reforming. Both of these situations in different blood proteins are discussed here.
Blood proteins regulated by allosteric disulfide bonds
We have chosen to highlight 4 soluble blood proteins and a lymphocyte receptor whose functions are controlled by cleavage of allosteric disulfide bonds. These examples are the best characterized at the molecular level so far. We also briefly discuss 7 other examples at a more preliminary stage of characterization.
Angiotensinogen
Plasma angiotensinogen is proteolyzed by renin and angiotensin-converting enzyme to produce the angiotensin peptides that control blood pressure and fluid homeostasis. The angiotensinogen Cys18–Cys138 disulfide bond is cleaved in a fraction of the plasma protein, which is significant because the oxidized protein is more efficiently activated by renin than the reduced protein.43 The ratio of oxidized to reduced angiotensinogen in blood is approximately 60:40 in healthy volunteers (independent of age or gender). This ratio increases in pregnant females with preeclampsia, which correlates with increased cleavage by renin and elevated blood pressure. It has been suggested that exposure of maternal angiotensinogen to reactive oxygen species present in the placenta increases the level of oxidized angiotensinogen, contributing to the development of preeclampsia.43
β2-glycoprotein I
Plasma β2-glycoprotein I is an autoantigen in the antiphospholipid syndrome, which is associated with vascular thrombosis, accelerated atherosclerosis, and recurrent miscarriages.44 The β2-glycoprotein I–antibody complex appears to prime the arterial and venous vasculature for thrombosis through effects on blood cells and coagulation and fibrinolysis proteins. β2-glycoprotein I consists of 4 complement modules and a unique fifth domain that contains a disulfide bond linking Cys288 and the C-terminal residue, Cys326.
The Cys288–Cys326 disulfide bond is cleaved in a fraction of plasma β2-glycoprotein I, and the bond is reduced in the purified protein by PDI and thioredoxin.34,35 The ratio of oxidized to reduced β2-glycoprotein I is elevated in the blood of patients with antiphospholipid syndrome compared with healthy volunteers and with patients with vascular thrombosis and no antiphospholipid antibodies.45 There is also more oxidized β2-glycoprotein I in the blood of patients with antiphospholipid syndrome with both anti–β2-glycoprotein I antibodies and lupus anticoagulant than patients with only anti–β2-glycoprotein I antibodies.45
The elevated oxidized β2-glycoprotein I in patients with antiphospholipid syndrome is associated with increased immunogenicity of the protein and increased thrombosis.45 Anti–β2-glycoprotein I antibodies from mice and rabbit plasma and autoantibodies from the plasma of patients with antiphospholipid syndrome bind more avidly to oxidized β2-glycoprotein I than reduced protein. The increased thrombosis may also relate to loss of anti-thrombotic properties of reduced β2-glycoprotein, such as protection of endothelial cells from oxidative stress.34
IL receptor subunit γ
Interleukin (IL) receptor subunit γ or CD132 is the common γ subunit of the cytokine receptors for IL-2, -4, -7, -9, -15, and -21 on the surface of lymphocytes. The CD132 Cys183–Cys232 disulfide bond is cleaved on the surface of cultured T cells by protein reductants and on the surface of thymocytes in mice after an inflammatory challenge.46 The disulfide bond is located at the subunit surface close to the IL-2 binding site.47 Reduction of the disulfide46 or replacement of Cys183 and Cys232 with alanine or serine48 inhibits IL-2 binding to the receptor complex and therefore receptor signaling and cell proliferation. Notably, Cys183 and Cys232 mutants are among those identified in patients with X-linked severe combined immunodeficiency.49,50
Plasminogen
Plasma plasminogen is the zymogen form of plasmin. The zymogen consists of an N-terminal Pan-apple domain followed by 5 kringle domains and a C-terminal serine protease domain. Plasminogen is converted to plasmin by urokinase or tissue plasminogen activator through cleavage of the Arg561–Val562 peptide bond in the serine protease domain and the Pan-apple domain is autoproteolytically released to produce mature plasmin. Plasmin activates other zymogens and cleaves the fibrin meshwork during dissolution of the thrombus and components of the extracellular matrix.
The Cys462–Cys541 disulfide bond in the kringle 5 domain is cleaved in a fraction of plasma plasminogen, and the level of reduced plasminogen varies in healthy individuals.41,51 Fragments of plasminogen containing the kringle domains are generated in plasma; these fragments function as endogenous inhibitors of tumor angiogenesis.52 The kringle fragments are produced from the reduced form of the zymogen.41 Generation of the kringle fragments from reduced plasminogen involves conversion to reduced plasmin, reduction of an additional kringle 5 disulfide bond, and subsequent proteolysis of up to 3 peptide bonds.41,53-55 Plasmin ligands such as tumor-derived phosphoglycerate kinase56 induce a conformational change in reduced kringle 5 that leads to attack by the Cys541 thiolate on the Cys536 sulfur atom of the Cys512–Cys536 disulfide bond, resulting in reduction of the bond by thiol-disulfide exchange (Figure 2B). Cleavage of the Cys512–Cys536 disulfide bond leads to a conformational change in plasmin and exposure of the peptide backbone to proteolysis on the C-terminal side of residues Lys486, Arg474, and Arg530. The proteolysis can be catalyzed by plasmin itself or other serine- or metalloproteases.
VWF
VWF is a plasma protein produced by vascular endothelial cells and megakaryocytes that chaperones blood coagulation cofactor factor VIII and tethers platelets to the injured blood vessel wall. It is a large glycoprotein that circulates as a series of multimers containing variable numbers of 500-kDa dimeric units. VWF structure is influenced by the mechanical shear forces in blood, where it transitions from a loosely coiled ball to elongated fibers that bind platelets.57-63
Fiber formation involves covalent self-association of VWF mediated by thiol-disulfide exchange in the VWF C2 domains.64,65 Both the C2 Cys2431–Cys2453 and nearby Cys2451–Cys2468 disulfide bonds are involved in fiber formation. Reduction of the C2 domain Cys2431–Cys2453 disulfide bond in 1 molecule, presumably by an oxidoreductase in blood, creates a Cys2431 thiolate anion that attacks the Cys2431 sulfur (of the Cys2431–Cys2453 disulfide bond) in another VWF molecule, resulting in a disulfide-linked dimer.42 The Cys2451–Cys2468 disulfide-dithiol then mediates formation of trimers and higher order oligomers.
Other blood proteins
Three blood cell receptors and 4 other soluble plasma proteins have been found to contain putative allosteric disulfides. These examples lack either in vivo evidence for a role for the bond and/or there is uncertainty as to the identity of the allosteric disulfide or disulfides in the protein. Their classification as allosteric disulfides should be considered preliminary at this stage.
Integrins are transmembrane, heterodimeric cell-adhesion receptors that mediate interactions between the cytoskeleton of cells and extracellular ligands and also play a role in signal transduction. Integrin activation has been associated with reduction of 1 or more disulfide bonds in the receptors.18,20,66-77 There are 18 α integrin subunits and 8 β subunits in mammals, but the majority of the redox studies have been conducted on the β3 subunit of the platelet αIIbβ3 (fibrinogen) receptor and the platelet and endothelial αvβ3 (vitronectin) receptor. There are 4 epidermal growth factor (EGF)-like domains in the extracellular region of β3.78,79 The disulfides in the EGF domains are generally important for maintaining the inactive state of β3 because mutation of a single cysteine for most of the disulfide bonds within the EGF domains results in a constitutively active β3.80-82 A recent study of the function of an atypical disulfide bond in β3 EGF domains is revealing.83 The disulfide bond in each of the 4 EGF domains in both αIIbβ3 and αvβ3 was investigated by mutating the cysteines to serines. An allosteric role for the EGF-4 Cys560–Cys583 disulfide in both αIIbβ3 and αvβ3 and for the EGF-3 Cys523–Cys544 bond only in αvβ3 was identified. For instance, activation of the C523S/C544S αvβ3 mutant by an antibody and a reducing agent is impaired. These disulfides are possible PDI19,20 and/or ERp5726,27 substrates.
TF is a transmembrane cofactor for factor VIIa. The TF/VIIa complex proteolytically activates factor X to initiate blood coagulation and provide the thrombin burst required for a stable thrombus. Intravascular TF is found on monocytes and neutrophils and exists mostly in a noncoagulant or cryptic form bound nonproductively to VIIa. Acute events lead to local decryption of TF, which appears to involve both formation of a disulfide bond between unpaired Cys186 and Cys209 in cryptic TF and exposure of phosphatidylserine on the cell surface (reviewed in Chen and Hogg84 ). Thioredoxin has been implicated in the maintenance of cryptic TF by cleaving the TF Cys186–Cys209 disulfide,84,85 whereas PDI has been implicated in TF decryption.86,87 The molecular mechanism of action of PDI in TF decryption is not known. Both the oxidoreductase and chaperone functions of PDI are possibly involved (reviewed in Langer and Ruf88 ).
Two circulating serine proteases appear to be regulated by allosteric disulfides. Proteolytic activation of coagulation factor XI contributes to the intrinsic/amplification phase of blood coagulation. One or more factor XI disulfide bonds are reduced in a fraction of the zymogen in blood, and the Cys362–Cys482 and Cys118–Cys147 disulfides are reduced by thioredoxin in the isolated enzyme.89 The reduced protein is more efficiently activated by thrombin, factor XIIa, or factor XIa than the oxidized protein, and blood levels of the reduced factor XI are elevated in patients with antiphospholipid syndrome,89 which may contribute to the thrombosis in this syndrome. Mast cell βII-tryptase is associated with pathological inflammation. The βII-tryptase Cys220–Cys248 disulfide bond exists in oxidized and reduced states in the secreted enzyme and the bond is reduced by thioredoxin in the isolated enzyme.36 The oxidized and reduced isoforms have different specificity and catalytic efficiency for hydrolysis of substrates.
The activity of 2 growth factors is also regulated by disulfide bond cleavage. Platelets contain abundant transforming growth factor-β1 (TGF-β1) that functions in a variety of physiologic and pathologic events. TGF-β1 is secreted as an inactive form in complex with latency-associated peptide and latent TGF-β–binding protein 1. Platelet TGF-β1 is activated by shear-induced disulfide bond cleavage in the growth factor,90 and platelet PDI has been implicated in this event.91 The identity of the functional disulfide bond or bonds in TGF-β1 is not known at present. The structures of the lymphangiogenic growth factors, vascular endothelial growth factor (VEGF)-C and VEGF-D, are also regulated by disulfide cleavage.92 The VEGFs function as disulfide-linked antiparallel homodimers, and dimer formation of VEGF-C and VEGF-D is controlled by a unique unpaired cysteine that exchanges with the interdimer disulfide bond.93
Detection of allosteric disulfide bonds in blood proteins
Allosteric disulfides are being identified using bioinformatics and experimental screens, and both approaches are proving useful for identifying these bonds in circulating proteins.
Bioinformatic approach
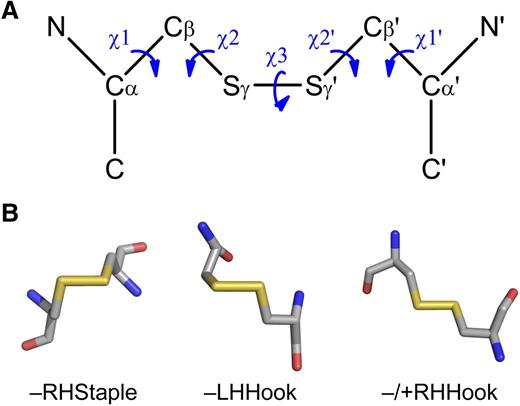
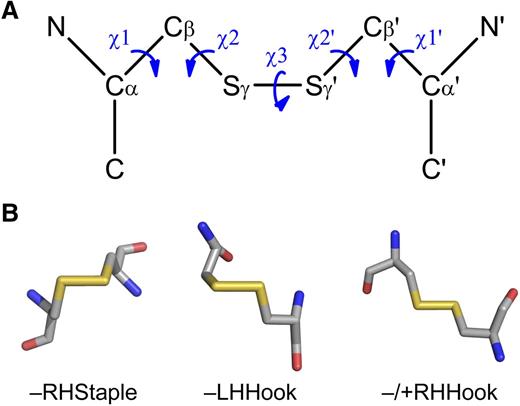
The bioinformatic technique relies on high-resolution 3-dimensional protein structures and a dataset of allosteric disulfides from which common features can be derived. The configuration, surface exposure, and secondary structural motifs that the disulfide links can be useful measures for the identification of allosteric bonds. These and other disulfide bond parameters are easily extracted from any protein structure in Protein Data Bank format.94 The most informative disulfide bond measure at this time is the configuration of the cystine residue. The geometry of cystine is defined by 5 dihedral or χ angles, which are calculated by the rotation around the bonds linking the 6 atoms (Figure 3A).95 There are 20 possible different cystine configurations, and all the types have been identified in protein crystal structures.95,96 Of the 20 different configurations, the right-handed (RH) –RHStaple, left-handed (LH) –LHHook, and −/+RHHook bonds are emerging as allosteric configurations (Figure 3B).2,8 The configurations of allosteric disulfide bonds in blood proteins and their mechanism of cleavage, where known, are summarized in Table 2.
Configurations of allosteric disulfide bonds. (A) Classification of disulfide bonds based on their geometry.95,96 The geometry is defined by the 5 bond angles (χ angles) linking the 2 α-carbons of the cystine residue. Cα, main chain carbon atom; Cβ, side chain carbon atom of each cysteine residue. The χ angles are recorded as being either positive or negative. The 3 basic types of bond configurations (spirals, hooks, and staples) are based on the signs of the central 3 angles, and they can be either RH or LH depending on whether the sign of the χ3 angle is positive or negative, respectively. These 6 bond types expand to 20 when the χ1 and χ1′ angles are taken into account. (B) Examples of the structures of the emerging allosteric configurations: –RHStaple, –LHHook, and −/+RHHook.
Configurations of allosteric disulfide bonds. (A) Classification of disulfide bonds based on their geometry.95,96 The geometry is defined by the 5 bond angles (χ angles) linking the 2 α-carbons of the cystine residue. Cα, main chain carbon atom; Cβ, side chain carbon atom of each cysteine residue. The χ angles are recorded as being either positive or negative. The 3 basic types of bond configurations (spirals, hooks, and staples) are based on the signs of the central 3 angles, and they can be either RH or LH depending on whether the sign of the χ3 angle is positive or negative, respectively. These 6 bond types expand to 20 when the χ1 and χ1′ angles are taken into account. (B) Examples of the structures of the emerging allosteric configurations: –RHStaple, –LHHook, and −/+RHHook.
It is important to keep in mind, though, that the protein backbone and the cystines that link it can change shape when in solution. That is, the crystal structure might represent only 1 of a number of different possible structures in solution. For instance, the same disulfide bond can exist in different configurations in different crystal structures.96 Dynamic isomerization of allosteric disulfide bonds is probably an important aspect of their function. Cleavage of the bond may only occur when the cystine adopts a particular configuration, which could be controlled by ligand binding or the mechanical shear in the circulation.
Experimental approach
A method based on differential cysteine labeling and mass spectrometry is currently the best experimental screen for allosteric disulfides.36,41,42,97 The approach is based on the principle that an allosteric disulfide will exist in equilibrium between reduced and oxidized states in a blood protein, such as observed in angiotensinogen, β2-glycoprotein I, and plasminogen. The technique measures the proportion of reduced and oxidized protein using different thiol alkylating agents. One alkylator is used to label the reduced portion of the allosteric disulfide bond in blood, or a fraction thereof, without or with previous treatment with a candidate oxidoreductase. The remaining disulfide bonds in the protein are then reduced using dithiothreitol and the new unpaired cysteines labeled with a second alkylator. Mass spectrometry is used to determine the identity of the protein and the ratio of differentially labeled cysteines. Typically, the cysteines showing significant increase in the ratio of the first to second alkylator after oxidoreductase treatment are allosteric disulfide candidates.
Summary
The functions of many blood proteins are controlled by cleavage of peptide bonds. For example, the blood coagulation and complement cascades are exquisitely regulated by discrete proteolysis. There are an increasing number of blood proteins found to be controlled by cleavage of the next most frequent covalent bond linking the polypeptide backbone of proteins: the disulfide bond. The allosteric and catalytic functional disulfide bonds contribute to the control of thrombosis and hemostasis, blood pressure, and inflammation in the circulation, and some bonds have been linked to disease. The pace of discovery of new allosteric disulfides in blood proteins is likely to increase during the coming years as new and better techniques for identifying and characterizing these bonds are developed. In terms of the oxidoreductases that cleave these bonds, the generation of specific inhibitors and cell-specific oxidoreductase-deficient mice has and will prove valuable for identifying the environments in which the enzymes work and their substrate disulfide bonds. Some allosteric and catalytic bonds will also likely be valid pharmaceutical targets and progress is being made on ways of targeting these bonds with small molecules or biologicals (reviewed in Hogg8 ).
Acknowledgments
This study was supported by grants from the National Health and Medical Research Council of Australia and the Cancer Council New South Wales.
Authorship
Contribution: P.J.H. conceived the review and all authors contributed to the writing of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Philip Hogg, Level 2, Lowy Cancer Research Centre, University of New South Wales, Sydney 2052, Australia; e-mail: p.hogg@unsw.edu.au.
References
Author notes
D.B. and K.M.C. contributed equally to this study.