Key Points
PSTPIP1 regulates the transition from podosomes to filopodia in macrophages by modulating WASP activity.
The novel PSTPIP1-R405C mutant induces filopodia formation, increases matrix degradation, and is associated with severe pyoderma gangrenosum.
Abstract
PSTPIP1 is a cytoskeletal adaptor and F-BAR protein that has been implicated in autoinflammatory disease, most notably in the PAPA syndrome: pyogenic sterile arthritis, pyoderma gangrenosum, and acne. However, the mechanism by which PSTPIP1 regulates the actin cytoskeleton and contributes to disease pathogenesis remains elusive. Here, we show that endogenous PSTPIP1 negatively regulates macrophage podosome organization and matrix degradation. We identify a novel PSTPIP1-R405C mutation in a patient presenting with aggressive pyoderma gangrenosum. Identification of this mutation reveals that PSTPIP1 regulates the balance of podosomes and filopodia in macrophages. The PSTPIP1-R405C mutation is in the SRC homology 3 (SH3) domain and impairs Wiskott-Aldrich syndrome protein (WASP) binding, but it does not affect interaction with protein-tyrosine phosphatase (PTP)-PEST. Accordingly, WASP inhibition reverses the elevated F-actin content, filopodia formation, and matrix degradation induced by PSTPIP1-R405C. Our results uncover a novel role for PSTPIP1 and WASP in orchestrating different types of actin-based protrusions. Our findings implicate the cytoskeletal regulatory functions of PSTPIP1 in the pathogenesis of pyoderma gangrenosum and suggest that the cytoskeleton is a rational target for therapeutic intervention in autoinflammatory disease.
Introduction
Dynamic regulation of the actin cytoskeleton and cell migration is essential for cellular immunity, because leukocytes travel long distances between tissues to perform their effector functions. Indeed, immunodeficiency syndromes, including Wiskott-Aldrich syndrome, leukocyte adhesion deficiency, and warts-hypogammaglobulinemia-infections-myelokathexis syndrome, are secondary to defects in the cytoskeleton and motility of leukocytes.1 Colchicine, which inhibits microtubule polymerization, is used to treat inflammatory conditions, and several other compounds that target cell motility are in development as immunomodulators, which indicates the importance of regulating the cytoskeleton to control immunity and inflammation.2,3 Conversely, neutrophils from patients with the autoinflammatory disease, neonatal onset multisystem inflammatory disease/Muckle-Wells syndrome, have impaired chemotaxis, which suggests that altered leukocyte migration may also promote a proinflammatory state.4 While altered leukocyte motility has been established as a cause of immunodeficiency, the role of cytoskeletal dysregulation and altered motility in inflammation and tissue damage remains poorly characterized.
Proline-serine-threonine phosphatase interacting protein 1 (PSTPIP1) is a cytoskeleton-associated adaptor and F-BAR domain–containing protein that is linked to PAPA syndrome, the human inflammatory disease consisting of pyogenic sterile arthritis, pyoderma gangrenosum, and acne.5,6 A hallmark of pyoderma gangrenosum is extensive tissue damage of unclear etiology.7 PAPA syndrome is considered to be an autoinflammatory disease because the adaptor function of PSTPIP1 links it to the inflammasome component, pyrin, to regulate interleukin-1β activity.8,9 However, patients with PAPA syndrome and pyoderma gangrenosum are often resistant to treatment with antiinflammatory agents that block tumor necrosis factor alpha and interleukin-1β signaling.10 Additionally, a mouse model of PAPA syndrome indicated that PSTPIP1 did not regulate the inflammasome in vivo, suggesting the interesting possibility that other PSTPIP1 adaptor functions are important in the pathogenesis of PAPA syndrome.11
The PSTPIP1 interacting partners, Wiskott-Aldrich syndrome protein (WASP) and protein-tyrosine phosphatase (PTP)-PEST (Protein Tyrosine Phosphatase, Non-Receptor Type 12 [PTPN12]), are important regulators of the cytoskeleton and cell migration, suggesting that PSTPIP1 mutations could contribute to disease through these pathways.12-15 WASP is a key activator of actin-related protein-2/3 (Arp2/3), and it plays an important role in nucleating new F-actin filaments and regulating F-actin–based structures in leukocytes.16 Macrophages deficient in WASP have impaired chemotaxis and lack podosomes, the actin-based adhesion structures of monocyte-lineage cells.13,17-19 Podosomes have a distinctive morphology consisting of a central “dot” of F-actin and F-actin polymerizing machinery surrounded by a “ring” of integrins and integrin-associated proteins.14,20 Podosomes also have extracellular matrix (ECM) degrading capabilities through the activity of matrix metalloproteinases.21,22 Two-dimensional (2D) macrophage chemotaxis seems to be dependent on podosomes, and it is believed that the degradative function of podosomes is important for three-dimensional (3D) invasion.13,23-25
Here, we show that endogenous PSTPIP1 negatively regulates podosome formation and matrix degradation. We also identified a novel PSTPIP1 mutation in a patient with aggressive pyoderma gangrenosum that revealed a role for PSTPIP1 in regulating a transition from podosome formation to filopodia formation and was correlated with increased matrix degradation. Our findings are the first to connect a disease-associated PSTPIP1 mutation to altered WASP activity and cytoskeletal organization. These findings provide the basis for new lines of investigation into autoinflammatory disease pathogenesis and novel therapeutic targets.
Materials and methods
Patient macrophages and PSTPIP1 sequencing
All research with human patients was performed in accordance with protocols approved by the University of Wisconsin-Madison Institutional Review Board. Verbal and written consent was obtained from all patients prior to participation in these studies. This study was conducted in accordance with the Declaration of Helsinki. Monocyte-derived macrophage isolation and culture were performed as previously described in detail.26 Differentiated macrophages were always used for experiments on day 7 after isolation.
For sequencing purposes, DNA was isolated from peripheral blood from the proband and both parents by using standard methods. The coding regions and splice sites of PSTPIP1 were resequenced by using both the forward and reverse directions on an automated ABI sequencer that used dye-terminator chemistry.
Gelatin degradation assays and immunofluorescence
THP-1 cells were acquired from American Type Culture Collection (ATCC) and grown according to ATCC guidelines. THP-1 cells were differentiated to a macrophage-like phenotype by treatment with phorbol 12-myristate 13-acetate as previously described by resuspending at 4 to 5 × 105 cells per milliliter in THP-1 medium supplemented with 12.5 ng/mL phorbol 12-myristate 13-acetate and culturing for 3 days prior to use.27 THP-1 cells were differentiated prior to use in all experiments.
Fluorescent gelatin coverslips were made by coating acid-washed coverslips with Oregon Green 488–conjugated gelatin (Invitrogen) or with Alexa Fluor 568–conjugated gelatin. Gelatin degradation assays were performed as previously described.26,28 For these assays, 5 × 104 cells were seeded on gelatin-coated coverslips for 3 hours. When used, wiskostatin and GM6001 were added for the entire duration of the assay. Degradation was quantified as previously described by using MetaMorph software.26 Briefly, a representative area was used to determine the average intensity of undegraded gelatin. The thresholding feature of the software was used to mark pixels with a fluorescence intensity value of 40% or less of the average intensity of undegraded gelatin. Immunofluorescence imaging of podosomes and filopodia was performed similarly to gelatin degradation imaging. In all, 5 × 104 macrophages or THP-1 cells were added to coverslips coated with 10 μg/mL fibrinogen and allowed to incubate for 3 to 4 hours (THP-1) or 24 hours (macrophages). When used, wiskostatin was added for the final 30 minutes of culture.
Chemotaxis and invasion assays
Chemotaxis assays were performed by using 6.5-mm Transwell inserts with 8-μm pores (Corning), and invasion assays were performed with matrigel-coated invasion chambers with 8-μm pores (BD) as previously described.29 Transwell inserts were then fixed and stained with a Hema III kit and quantified by counting the number of cells in 5 separate ×40 fields for each insert. Invasion assays were quantified by acquiring 3 separate grayscale images of each insert on a Nikon SMZ1500 stereomicroscope at ×11.25 optical zoom. The thresholding feature in MetaMorph was used to mark the cell-containing areas. The color camera on this microscope was used to take representative images of Transwell and invasion chamber results.
Generation of PSTPIP1 knockdown and rescue THP-1 cell lines
Two sets of PSTPIP1 knockdown lines were made for this study. We made PSTPIP1 and control knockdown cells by using lentiviral short hairpin RNA (shRNA) constructs prepackaged in lentiviral particles (sc-90246-V and sc-108080; Santa Cruz Biotechnology) and lentivirus made by using individual targets in pLKO.1 vector purchased from Open Biosystems. Infections were performed by pelleting 106 log-phase THP-1 cells and resuspending them in medium containing viral particles and 4 μg/mL polybrene according to previously described methods.26,30 PSTPIP1 rescue cell lines were made by retroviral transduction of control or PSTPIP1 (TRCN0000002988) knockdown cell lines with pMX-IRES-GFP or pMX-GFP-PSTPIP1-WT or -R405C. Three days later, fluorescence-activated cell sorting (FACS) was used to separate green fluorescent protein (GFP)-positive from GFP-negative cells. The GFP-positive population was expanded and subjected to a second round of FACS to isolate a population with low GFP/GFP-PSTPIP1 expression.
Protein purification and in vitro pulldown assays
Glutathione-S-transferase (GST)-tagged GST-PSTPIP1–wild-type (WT), –R405C, and –ΔSH3 (SRC homology 3) were purified from E coli lysates with Glutathione Uniflow resin (Clontech), and His-WASP was purified from E coli lysates by using nickel-nitrilotriacetic acid superflow resin (Qiagen). For in vitro interaction studies, 4 μL of GST-PSTPIP beads and 40 μL of His-WASP were mixed and incubated for 4 hours at 4°C and washed 3 times with the assay buffer. Western blotting was used to determine the degree of interaction.
F-actin content assays
F-actin content assays were performed as previously described.31 The wells of a 24-well plate were coated with 10 μg/mL fibrinogen, and 2.5 × 105 THP-1 cells were added and differentiated for 2.5 days. The cells were serum starved in medium containing 0.2% bovine serum albumin for 12 hours and stimulated for 5 minutes in complete medium or with or without 100 ng/mL chemokine (C-C motif) ligand 2. F-actin was saturated with 1.5 μM rhodamine phalloidin in phosphate-buffered saline for 10 minutes at room temperature, and background fluorescence was determined by adding 150 μM unlabeled phalloidin during staining. Phalloidin was extracted with 100% methanol and transferred to a 96-well assay plate; fluorescence intensity was read on a Victor3 V plate reader with Excitation 531 nm and Emission 590/10 nm filters (Perkin Elmer).
Statistical analysis
Statistical analysis was performed by using GraphPad Prism software. Significance was determined by using paired or unpaired Student t tests and one-way analysis of variance (ANOVA) as indicated in the figure legends. A difference was considered significant at P < .05. All statistical analyses were performed on data from at least 3 independent experiments.
Results
PSTPIP1 impairs podosome formation and function in THP-1 macrophages
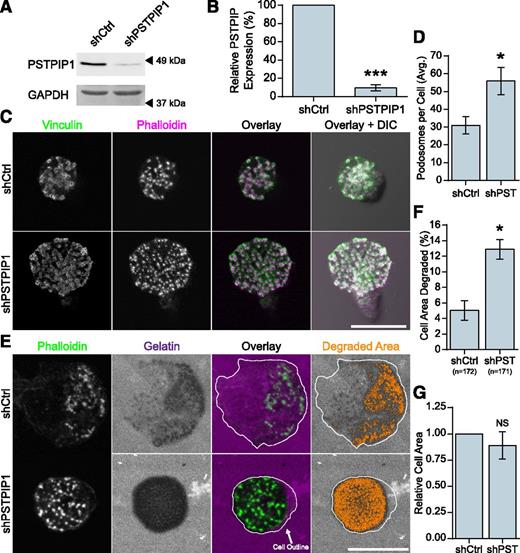
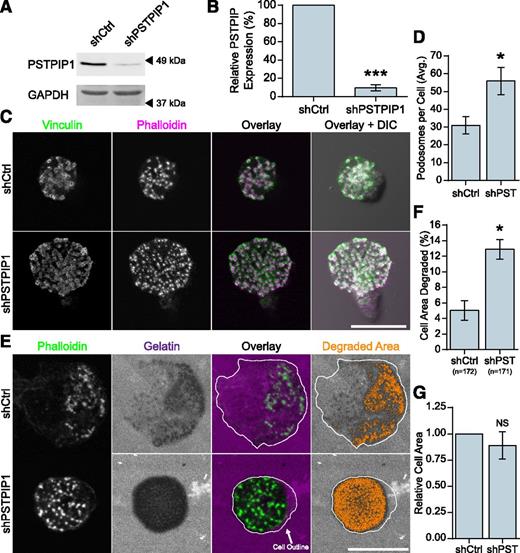
Previous work has demonstrated a role for the PSTPIP1 paralog, PSTPIP2, which is related to PSTPIP1 but lacks an SH3 domain and WASP binding, in podosome maturation and filopodia formation.32,33 To determine whether endogenous PSTPIP1 regulates macrophage podosomes, we performed shRNA-mediated depletion of PSTPIP1 in THP-1 macrophage-like cells. We achieved a 90% knockdown of PSTPIP1 protein (Figure 1A-B), and the following results were confirmed by using a set of pooled PSTPIP1 shRNA targets (supplemental Figure 1A-F on the Blood Web site). PSTPIP1-deficient cells formed significantly more podosomes compared with control cells (Figure 1C-D). Because ECM degradation is an important function of podosomes, we tested the degradative ability of PSTPIP1-deficient cells by using a fluorescent gelatin assay. PSTPIP1-deficient cells degraded more fluorescent gelatin than controls (Figure 1E-F), and there was no difference in the spread area between the two cell lines (Figure 1G). Taken together, these findings suggest that endogenous PSTPIP1 negatively regulates podosome formation and ECM degradation in macrophages.
PSTPIP1 impairs podosome formation. (A-B) Lentiviral transduction of shRNA was used to knock down PSTPIP1 in THP-1 cells. (A) Western blot of lysates from the control (shCtrl) and PSTPIP1 knockdown (shPSTPIP1 or shPST) THP-1 cells using anti-PSTPIP1 and anti–glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibodies (loading control). Arrowheads indicate location of molecular weight markers. (B) Quantification of PSTPIP1 knockdown in which PSTPIP1 expression in shCtrl cells was normalized to 100%. (C) Representative images of podosomes in shCtrl and shPSTPIP1 THP-1 macrophages. Podosomes were stained with anti-vinculin antibody (green) and rhodamine phalloidin (magenta). (D) Quantification of the average number of podosomes formed by shCtrl and shPSTPIP1 THP-1 cells. (E) Representative images of gelatin degradation by PSTPIP1 knockdown cells on Alexa Fluor 568–conjugated gelatin (magenta) coverslips. Podosomes were stained with Alexa Fluor 488 phalloidin (green). The cell outline is a trace of the cell border made from the phalloidin image. Orange pixels in the Degraded Area panel were those counted as degraded. (F) Quantification of the amount of gelatin degraded by PSTPIP1 knockdown cells as a percentage of cell area. (G) Cell area was determined in all cells for the degradation experiment and normalized to the shCtrl value. Scale bars, 20 μm. All values are mean ± standard error of the mean (SEM) from 3 (D,F,G) or 4 (B) independent experiments. *P < .05; ***P < .001 as determined by a paired Student t test (B,D,G) or ANOVA with repeated measures using a compound symmetry correlation structure (F). NS, nonsignificant.
PSTPIP1 impairs podosome formation. (A-B) Lentiviral transduction of shRNA was used to knock down PSTPIP1 in THP-1 cells. (A) Western blot of lysates from the control (shCtrl) and PSTPIP1 knockdown (shPSTPIP1 or shPST) THP-1 cells using anti-PSTPIP1 and anti–glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibodies (loading control). Arrowheads indicate location of molecular weight markers. (B) Quantification of PSTPIP1 knockdown in which PSTPIP1 expression in shCtrl cells was normalized to 100%. (C) Representative images of podosomes in shCtrl and shPSTPIP1 THP-1 macrophages. Podosomes were stained with anti-vinculin antibody (green) and rhodamine phalloidin (magenta). (D) Quantification of the average number of podosomes formed by shCtrl and shPSTPIP1 THP-1 cells. (E) Representative images of gelatin degradation by PSTPIP1 knockdown cells on Alexa Fluor 568–conjugated gelatin (magenta) coverslips. Podosomes were stained with Alexa Fluor 488 phalloidin (green). The cell outline is a trace of the cell border made from the phalloidin image. Orange pixels in the Degraded Area panel were those counted as degraded. (F) Quantification of the amount of gelatin degraded by PSTPIP1 knockdown cells as a percentage of cell area. (G) Cell area was determined in all cells for the degradation experiment and normalized to the shCtrl value. Scale bars, 20 μm. All values are mean ± standard error of the mean (SEM) from 3 (D,F,G) or 4 (B) independent experiments. *P < .05; ***P < .001 as determined by a paired Student t test (B,D,G) or ANOVA with repeated measures using a compound symmetry correlation structure (F). NS, nonsignificant.
A novel PAPA syndrome–associated mutation
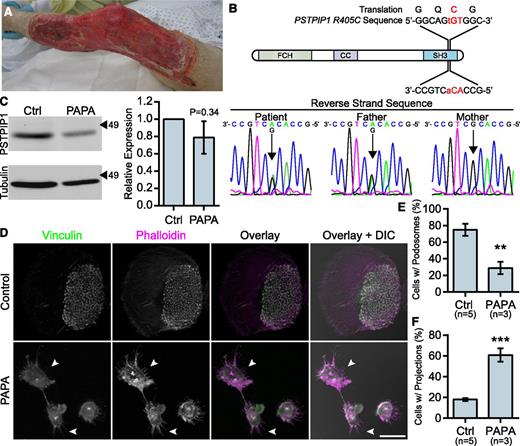
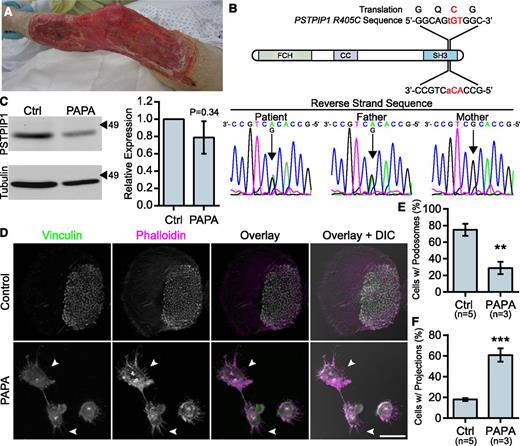
To better understand PSTPIP1 function in the context of human disease, we characterized macrophages from a patient with a novel PSTPIP1 mutation. The patient presented with recurrent episodes of severe pyoderma gangrenosum without a history of arthritis. His father had a history of severe acne. At presentation with an episode of new onset pyoderma gangrenosum, he was found to have elevated inflammatory markers and normal integrin (CD11b/CD18) surface expression (Figure 2A and supplemental Figure 2A-B). Sequencing revealed that the patient and his father were heterozygous for a c.1213C>T substitution in exon 15 of PSTPIP1, resulting in a p.Arg405Cys substitution (PSTPIP1-R405C) at the amino acid level (Figure 2B). This variation has been reported in 6 of 14 716 chromosomes in the 1000 Genomes Browser and Exome Variant Server, and the sorting intolerant from tolerant algorithm predicted that it would have a deleterious effect on protein function.34-37 Immunoblotting revealed a slight, but nonsignificant decrease in PSTPIP1 expression in monocyte-derived macrophages, suggesting that PSTPIP1 stability was not substantially altered (Figure 2C). Previously reported PSTPIP1 mutations associated with PAPA syndrome have been described between amino acids 230 and 266, an area which lies between the N-terminal F-BAR domain and the C-terminal SH3 domain (supplemental Figure 2C).6,10,38-41 This is the first PAPA-associated mutation that has been found in the SH3 domain of PSTPIP1, which mediates interactions with WASP, pyrin, Abelson murine leukemia viral oncogene homolog 1 (ABL-1; c-Abl), and Fas ligand.8,42-44
Macrophages from a PAPA syndrome patient have increased filopodia-like membrane projections. (A) Image of pyoderma gangrenosum on the PAPA patient’s right leg. (B) Reverse strand sequence traces from the PSTPIP1 genomic DNA sequencing of the patient, his mother, and his father. The diagram shows the location of the mutation at the DNA and protein level. (C) Western blots of lysate from control and PAPA macrophages was probed with mouse anti-PSTPIP1 and rabbit anti-β-tubulin (loading control) antibodies to determine relative PSTPIP1 expression. Arrowheads indicate location of molecular weight markers (kDa). PSTPIP1 expression in the PAPA patient macrophages was normalized to controls for quantification. (D) Representative images of podosomes and filopodia-like membrane projections in PAPA and control macrophages. Podosomes and filopodia-like structures were stained with anti-vinculin antibody (green) and rhodamine phalloidin (magenta). Arrowheads indicate filopodia-like projections in PAPA macrophages. (E-F) Quantification of the percentage of macrophages shown in (D) that form podosomes (E) and filopodia-like projections (F). Scale bar, 20 μm. All values are mean ± SEM from 3 (E,F) or 4 (C) independent experiments. **P < .01; ***P < .001 as determined by an unpaired Student t test (E,F) or paired Student t test (C).
Macrophages from a PAPA syndrome patient have increased filopodia-like membrane projections. (A) Image of pyoderma gangrenosum on the PAPA patient’s right leg. (B) Reverse strand sequence traces from the PSTPIP1 genomic DNA sequencing of the patient, his mother, and his father. The diagram shows the location of the mutation at the DNA and protein level. (C) Western blots of lysate from control and PAPA macrophages was probed with mouse anti-PSTPIP1 and rabbit anti-β-tubulin (loading control) antibodies to determine relative PSTPIP1 expression. Arrowheads indicate location of molecular weight markers (kDa). PSTPIP1 expression in the PAPA patient macrophages was normalized to controls for quantification. (D) Representative images of podosomes and filopodia-like membrane projections in PAPA and control macrophages. Podosomes and filopodia-like structures were stained with anti-vinculin antibody (green) and rhodamine phalloidin (magenta). Arrowheads indicate filopodia-like projections in PAPA macrophages. (E-F) Quantification of the percentage of macrophages shown in (D) that form podosomes (E) and filopodia-like projections (F). Scale bar, 20 μm. All values are mean ± SEM from 3 (E,F) or 4 (C) independent experiments. **P < .01; ***P < .001 as determined by an unpaired Student t test (E,F) or paired Student t test (C).
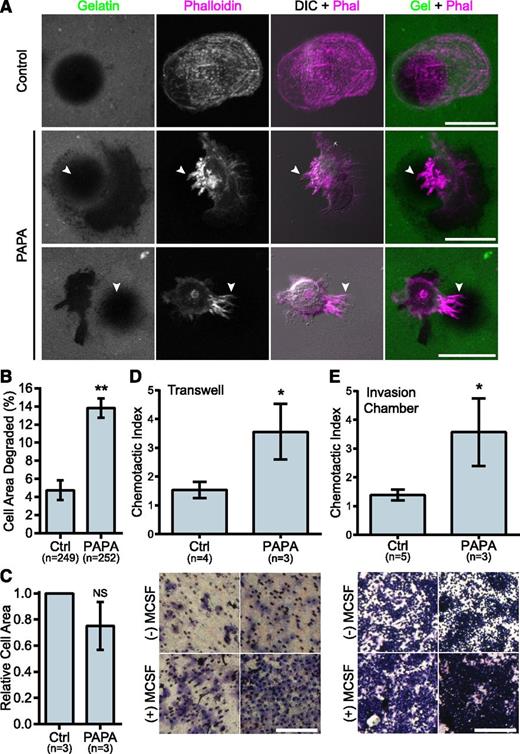
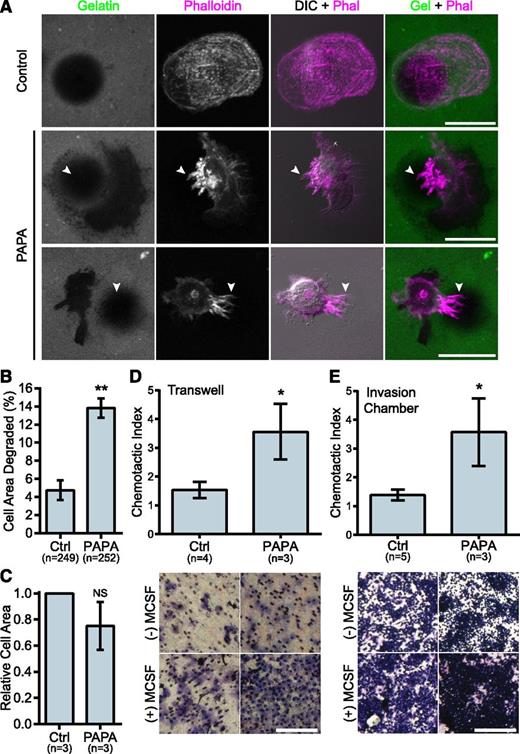
We previously demonstrated that macrophages from PAPA patients with a p.Ala230Thr mutation formed fewer podosomes, had impaired matrix degradation, and were less migratory than controls, suggesting a gain-of-function mutation.29 Surprisingly, in macrophages from the patient with the R405C mutation, we found few podosomes but robust matrix degradation associated with filopodia in 2D environments. Control donor macrophages showed typical podosomes characterized by F-actin cores surrounded by vinculin rings that organized into rosettes. However, the patient’s macrophages had very few podosomes and had instead large clusters of F-actin that did not colocalize with vinculin (Figure 2D-E and supplemental Figure 2D-F). Strikingly, the patient’s macrophages formed prominent membrane projections resembling filopodia that were rare in control donors (Figure 2F). The control macrophages degraded patches of gelatin that were directly underneath the cell and that were often contained under clusters of podosomes. However, the PAPA patient macrophages were highly degradative, despite their lack of podosomes. We frequently observed that the degraded gelatin was distributed around the cell, rather than underneath it, and that the degraded areas were located under clusters of membrane projections that extended from the cells (Figure 3A-B). We also found that the patient macrophages appeared to be less spread; however, there was not a significant difference in cell area between PAPA patient macrophages and control (Figure 3C). Thus, our data suggested that PAPA patient macrophages may be able to degrade ECM through filopodia-like protrusions rather than podosomes. Podosome formation has been correlated with the ability of macrophages to perform chemotaxis and invasive migration.25,45 However, we found that the current patient macrophages, in contrast to macrophages from other PAPA patients, were more chemotactic and invasive compared with controls (Figure 3D-E).29
PAPA patient macrophages make membrane projections associated with matrix degradation and have enhanced invasive properties. (A) Representative images of gelatin degradation by control and PAPA macrophages on Oregon Green 488 gelatin-coated coverslips (green). Podosomes and membrane projections were stained with rhodamine phalloidin (magenta). Areas degraded by the macrophages appear black in the gelatin image. Arrowheads indicate projections overlaying degraded areas. (B) Quantification of gelatin degraded by control and PAPA macrophages as a percentage of the total cell area. (C) Cell area was determined for all cells in the assay and normalized to the value of the control macrophages. (D-E) Quantification of macrophage chemotaxis to macrophage colony-stimulating factor (M-CSF) in (D) Transwell devices or (E) Matrigel-coated invasion chambers. Chemotactic index is the ratio of cells migrating to M-CSF over medium alone. Representative images of the filters are shown below. Scale bars, 20 μm (A) or 500 μm (D,E). All values are mean ± SEM from 3 independent experiments. *P < .05; **P < .01 as determined by a paired Student t test (C) unpaired Student t test (D,E) or ANOVA with repeated measures using a compound symmetry correlation structure (B).
PAPA patient macrophages make membrane projections associated with matrix degradation and have enhanced invasive properties. (A) Representative images of gelatin degradation by control and PAPA macrophages on Oregon Green 488 gelatin-coated coverslips (green). Podosomes and membrane projections were stained with rhodamine phalloidin (magenta). Areas degraded by the macrophages appear black in the gelatin image. Arrowheads indicate projections overlaying degraded areas. (B) Quantification of gelatin degraded by control and PAPA macrophages as a percentage of the total cell area. (C) Cell area was determined for all cells in the assay and normalized to the value of the control macrophages. (D-E) Quantification of macrophage chemotaxis to macrophage colony-stimulating factor (M-CSF) in (D) Transwell devices or (E) Matrigel-coated invasion chambers. Chemotactic index is the ratio of cells migrating to M-CSF over medium alone. Representative images of the filters are shown below. Scale bars, 20 μm (A) or 500 μm (D,E). All values are mean ± SEM from 3 independent experiments. *P < .05; **P < .01 as determined by a paired Student t test (C) unpaired Student t test (D,E) or ANOVA with repeated measures using a compound symmetry correlation structure (B).
PSTPIP1-R405C impairs podosome formation and promotes filopodia formation
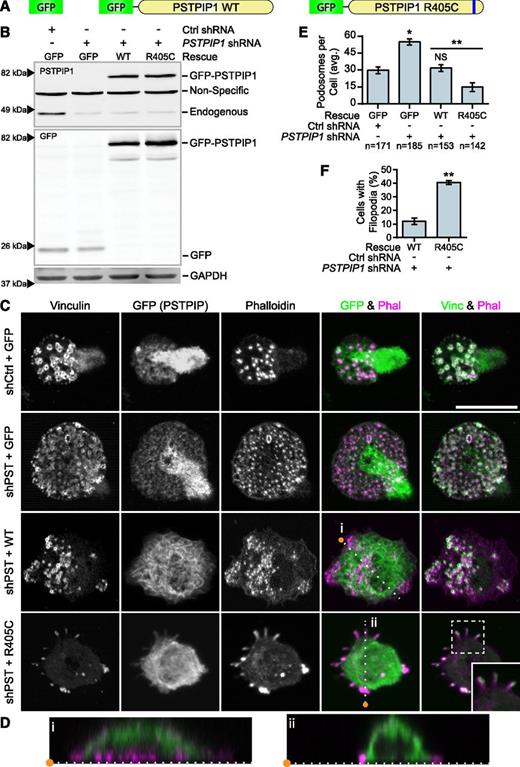
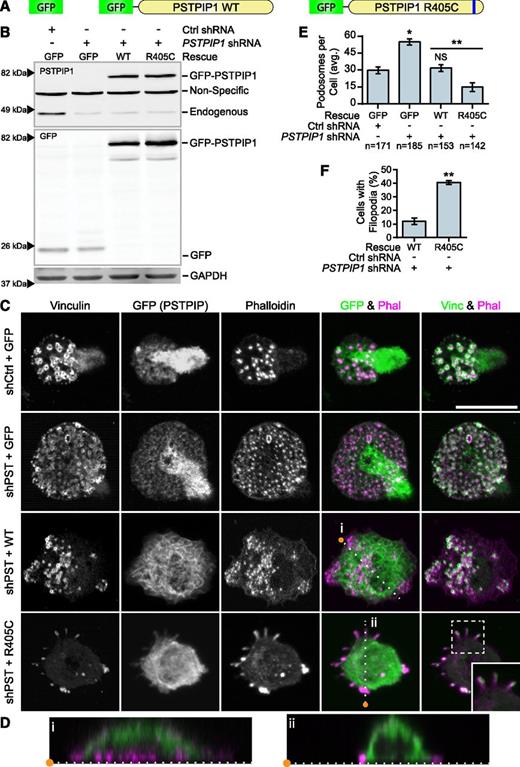
To determine whether the PSTPIP1-R405C mutation was sufficient to cell-autonomously recapitulate the PAPA phenotype in vitro, we rescued the PSTPIP1-deficient THP-1 cells with either PSTPIP1-WT or PSTPIP1-R405C (Figure 4A). We used FACS to acquire a population of cells with expression near endogenous levels of PSTPIP1 (Figure 4B and supplemental Figure 3). Cell lines expressing GFP alone had fluorescence evenly distributed throughout the cell body. PSTPIP1-WT and -R405C localized to the dorsal plasma membrane and could induce membrane tubulation, as previously reported (Figure 4C-D).46,47 Interestingly, and in contrast to PSTPIP2, PSTPIP1 did not localize to podosomes.32 As expected, PSTPIP1-deficient cells expressing GFP had approximately double the number of podosomes compared with control cells expressing GFP. Rescue with PSTPIP1-WT restored the podosome numbers to the level of the control. By contrast, expression of PSTPIP1-R405C impaired podosome formation, similar to macrophages from our patient (Figure 4C,E). PSTPIP1-R405C–expressing cells formed significantly more membrane projections compared with cells expressing PSTPIP1-WT, confirming the findings from our patient (Figure 4C,F). To determine whether these membrane protrusions were filopodia-like, we stained cells expressing PSTPIP1-WT and PSTPIP1-R405C for the filopodia markers, Vasodilator-stimulated phosphoprotein (VASP) and Arp2/3. We found that the membrane extensions were enriched with both VASP and Arp2/3, indicating that PSTPIP1-R405C induces filopodia formation (Figure 5A-B and supplemental Figure 4A-B). Moreover, live differential interference contrast imaging showed that these structures were protrusive rather than retraction fibers (data not shown). Taken together, the findings demonstrate that the PSTPIP1-R405C mutation is sufficient to induce a switch from podosomes to the formation of filopodia.
PSTPIP1-R405C promotes filopodia formation. (A) Diagram of constructs introduced into control and PSTPIP1 knockdown cells by retroviral transduction to make rescue cell lines. (B) Representative western blot of lysates from FACS-sorted THP-1 rescue lines showing expression of rescue constructs. Blots were probed with mouse anti-PSTPIP1, rabbit anti-GFP, and mouse anti-GAPDH (loading control) antibodies. Arrowheads indicate location of molecular weight markers. (C) Representative images of podosomes and filopodia formed by the THP-1 rescue cell lines, which were stained with anti-vinculin antibody (green) and rhodamine phalloidin (magenta). Anti-GFP antibody (green) was used to amplify the GFP signal. Inset shows filopodia at ×1.5 original magnification. (D) Slices through the Z-plane of the shPST + WT and shPST + R405C cells were created with the Reslice function in FIJI software as indicated by the dashed lines in (C). (E) Quantification of the average number of podosomes formed by the indicated rescue line. shCtrl + GFP was the reference value for statistical comparison except where indicated by a bar. (F) Quantification of the percentage of shPST + WT and shPST + R405C cells that form filopodia. Scale bars, 20 μm. All values are mean ± SEM from 3 independent experiments. *P < .05; **P < .01 as determined by ANOVA with repeated measures using a compound symmetry correlation structure (E) or paired Student t test (F). NS, nonsignificant.
PSTPIP1-R405C promotes filopodia formation. (A) Diagram of constructs introduced into control and PSTPIP1 knockdown cells by retroviral transduction to make rescue cell lines. (B) Representative western blot of lysates from FACS-sorted THP-1 rescue lines showing expression of rescue constructs. Blots were probed with mouse anti-PSTPIP1, rabbit anti-GFP, and mouse anti-GAPDH (loading control) antibodies. Arrowheads indicate location of molecular weight markers. (C) Representative images of podosomes and filopodia formed by the THP-1 rescue cell lines, which were stained with anti-vinculin antibody (green) and rhodamine phalloidin (magenta). Anti-GFP antibody (green) was used to amplify the GFP signal. Inset shows filopodia at ×1.5 original magnification. (D) Slices through the Z-plane of the shPST + WT and shPST + R405C cells were created with the Reslice function in FIJI software as indicated by the dashed lines in (C). (E) Quantification of the average number of podosomes formed by the indicated rescue line. shCtrl + GFP was the reference value for statistical comparison except where indicated by a bar. (F) Quantification of the percentage of shPST + WT and shPST + R405C cells that form filopodia. Scale bars, 20 μm. All values are mean ± SEM from 3 independent experiments. *P < .05; **P < .01 as determined by ANOVA with repeated measures using a compound symmetry correlation structure (E) or paired Student t test (F). NS, nonsignificant.
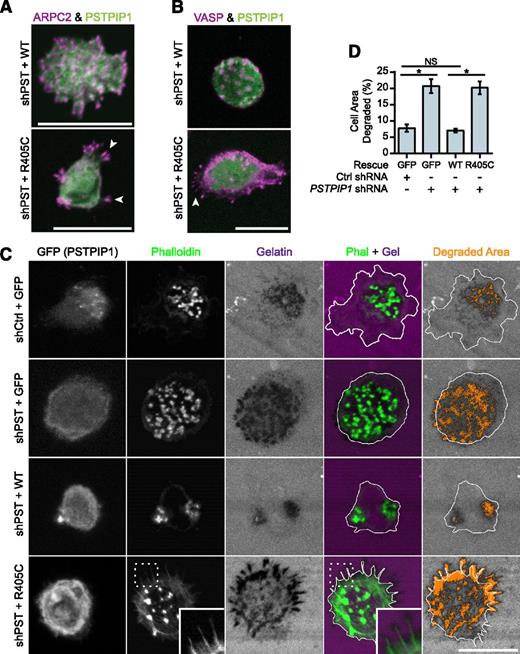
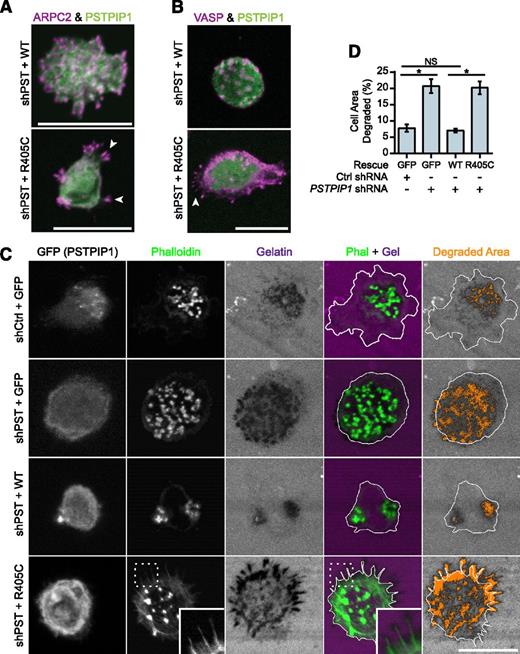
Filopodia associated with matrix degradation are induced by PSTPIP1 R405C. (A-B) Representative images of podosomes and filopodia formed in the THP-1 rescue cell lines. (A) The Arp2/3 complex was stained in podosomes and filopodia using anti-ARPC2 antibody (magenta) in THP-1 rescue cells expressing GFP-tagged PSTPIP1 constructs (green). (B) VASP was labeled in podosomes and filopodia with anti-VASP antibody (magenta) in THP-1 rescue cells expressing GFP-tagged PSTPIP1 constructs (green). (C) Representative images of gelatin degradation in the THP-1 rescue lines plated on Alexa Fluor 568–conjugated gelatin-coated coverslips (magenta). F-actin was stained with CruzFluor 405 phalloidin (green), and anti-GFP antibody (gray) was used to amplify the GFP signal. Orange pixels in the Degraded Area panel were those counted as degraded. (D) Quantification of the amount of gelatin degraded by the THP-1 rescue cells as a percentage of cell area. Scale bar, 20 μm. Values are mean ± SEM from 4 independent experiments. *P < .05, as determined by one-way ANOVA with Sidak’s multiple comparisons test. NS, nonsignificant.
Filopodia associated with matrix degradation are induced by PSTPIP1 R405C. (A-B) Representative images of podosomes and filopodia formed in the THP-1 rescue cell lines. (A) The Arp2/3 complex was stained in podosomes and filopodia using anti-ARPC2 antibody (magenta) in THP-1 rescue cells expressing GFP-tagged PSTPIP1 constructs (green). (B) VASP was labeled in podosomes and filopodia with anti-VASP antibody (magenta) in THP-1 rescue cells expressing GFP-tagged PSTPIP1 constructs (green). (C) Representative images of gelatin degradation in the THP-1 rescue lines plated on Alexa Fluor 568–conjugated gelatin-coated coverslips (magenta). F-actin was stained with CruzFluor 405 phalloidin (green), and anti-GFP antibody (gray) was used to amplify the GFP signal. Orange pixels in the Degraded Area panel were those counted as degraded. (D) Quantification of the amount of gelatin degraded by the THP-1 rescue cells as a percentage of cell area. Scale bar, 20 μm. Values are mean ± SEM from 4 independent experiments. *P < .05, as determined by one-way ANOVA with Sidak’s multiple comparisons test. NS, nonsignificant.
PSTPIP1-R405C promotes ECM degradation
To determine whether PSTPIP1-R405C was sufficient to induce elevated ECM degradation, we performed fluorescent gelatin degradation assays. The PSTPIP1-deficient cells (+GFP) degraded approximately threefold more gelatin than the control (+GFP) cells (Figures 5C-D and 1E-F). Addition of PSTPIP1-WT rescued the gelatin degradation to control levels. However, PSTPIP1-R405C did not rescue the gelatin degradation, and the cells showed a threefold increase in gelatin degradation compared with cells expressing comparable levels of PSTPIP1-WT (Figure 5C-D). Although the R405C-expressing cells did not form large clusters of filopodia over areas of degradation as seen with the PAPA macrophages (Figure 3A), degradation was localized beneath individual filopodia (Figure 5C). Thus, PSTPIP1-R405C impairs podosomes and induces a switch to filopodia that is associated with increased matrix degradation.
PSTPIP1-R405C has decreased binding to WASP
Because PSTPIP1 binds WASP through its SH3 domain, we sought to determine whether the R405C mutation affected its interaction with WASP.42,48 Additionally, PSTPIP1 is able to mediate the interaction between the tyrosine phosphatase PTP-PEST and WASP, which promotes dephosphorylation of WASP Y291.48 In F-actin polymerization assays, PSTPIP1 and PTP-PEST decrease WASP-dependent actin polymerization.49 Therefore, we sought to determine whether the R405C mutation altered interactions with WASP or PTP-PEST. We performed coimmunoprecipitation assays using the THP-1 rescue lines to determine whether the R405C mutation affected WASP binding. We found that WASP immunoprecipitated with PSTPIP1-WT but only at background levels with PSTPIP1-R405C (Figure 6A). To further confirm that the R405C mutation disrupts WASP binding, we purified WASP and PSTPIP1 mutants from E coli for use in pulldown assays (Figure 6B). We found that the interaction of His-WASP with bead-bound GST-PSTPIP1-R405C was reduced by approximately 50% compared with PSTPIP1-WT and that deleting the SH3 domain completely abrogated the interaction (Figure 6C). Both WASP and neural WASP (N-WASP) are expressed in macrophages, although WASP is expressed at much higher levels.50 By using coimmunoprecipitation, we found that PSTPIP1 can interact with N-WASP in addition to WASP (supplemental Figure 5A).
PSTPIP1-R405C has impaired interaction with WASP but not PTP-PEST. (A) Anti-GFP antibody was used to immunoprecipitate GFP or GFP-tagged PSTPIP1 constructs from the indicated THP-1 rescue cell lines. Blots were probed with anti-GFP antibody and anti-WASP antibody to visualize the coimmunoprecipitation (IP) of WASP. Equivalent loading was determined by probing a blot of the lysates as above. (B) His-tagged WASP or GST-tagged PSTPIP1 proteins (labeled as GST-WT, GST-R405C, and GST-ΔSH3) were purified from E coli. They were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie brilliant blue to assess purity. (C) Pulldown assay using the purified proteins described in (B). Soluble His-WASP was incubated with the indicated bead-bound GST-PSTPIP1 construct. Blots were probed for WASP and GST (to detect PSTPIP1) to determine the degree of interaction. The amount of WASP pulled down by the GST-PSTPIP1 mutants was normalized to the interaction with GST-PSTPIP1-WT. (D) Anti-GFP antibody was used to coimmunoprecipitate PTP-PEST with GFP or GFP-tagged PSTPIP1 constructs from the indicated THP-1 rescue cell lines. Anti–PTP-PEST and anti-GFP antibodies were used to probe the immunoprecipitation and input blots. (E) Quantification of the amount of PTP-PEST coimmunoprecipitating with PSTPIP1 was determined as a percent of input and normalized to the PSTPIP1-WT value. (F) Anti-GFP antibody was used to immunoprecipitate GFP-PSTPIP1-WT or -R405C from THP-1 rescue cells. Immunoprecipitates and lysates were probed with anti-phosphotyrosine (4G10 platinum) and anti-GFP antibodies to assess PSTPIP1 phosphorylation. (G) Quantification of the phosphorylation of PSTPIP1-WT and -R405C is shown relative to WT. Unlabeled arrowheads indicate the band(s) of interest in each panel. Labeled arrowheads indicate the locations of the molecular weight markers (kDa). All values are mean ± SEM from 3 (C,G) or 4 (E) independent experiments. **P < .01; ****P < .0001 as determined by one-way ANOVA with Sidak’s multiple comparisons test (C,E) or paired Student t test (G). IB, immunoblotting. NS, nonsignificant.
PSTPIP1-R405C has impaired interaction with WASP but not PTP-PEST. (A) Anti-GFP antibody was used to immunoprecipitate GFP or GFP-tagged PSTPIP1 constructs from the indicated THP-1 rescue cell lines. Blots were probed with anti-GFP antibody and anti-WASP antibody to visualize the coimmunoprecipitation (IP) of WASP. Equivalent loading was determined by probing a blot of the lysates as above. (B) His-tagged WASP or GST-tagged PSTPIP1 proteins (labeled as GST-WT, GST-R405C, and GST-ΔSH3) were purified from E coli. They were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie brilliant blue to assess purity. (C) Pulldown assay using the purified proteins described in (B). Soluble His-WASP was incubated with the indicated bead-bound GST-PSTPIP1 construct. Blots were probed for WASP and GST (to detect PSTPIP1) to determine the degree of interaction. The amount of WASP pulled down by the GST-PSTPIP1 mutants was normalized to the interaction with GST-PSTPIP1-WT. (D) Anti-GFP antibody was used to coimmunoprecipitate PTP-PEST with GFP or GFP-tagged PSTPIP1 constructs from the indicated THP-1 rescue cell lines. Anti–PTP-PEST and anti-GFP antibodies were used to probe the immunoprecipitation and input blots. (E) Quantification of the amount of PTP-PEST coimmunoprecipitating with PSTPIP1 was determined as a percent of input and normalized to the PSTPIP1-WT value. (F) Anti-GFP antibody was used to immunoprecipitate GFP-PSTPIP1-WT or -R405C from THP-1 rescue cells. Immunoprecipitates and lysates were probed with anti-phosphotyrosine (4G10 platinum) and anti-GFP antibodies to assess PSTPIP1 phosphorylation. (G) Quantification of the phosphorylation of PSTPIP1-WT and -R405C is shown relative to WT. Unlabeled arrowheads indicate the band(s) of interest in each panel. Labeled arrowheads indicate the locations of the molecular weight markers (kDa). All values are mean ± SEM from 3 (C,G) or 4 (E) independent experiments. **P < .01; ****P < .0001 as determined by one-way ANOVA with Sidak’s multiple comparisons test (C,E) or paired Student t test (G). IB, immunoblotting. NS, nonsignificant.
Coimmunoprecipitation assays were also performed to test the interaction between PSTPIP1 and PTP-PEST. By using lysates from the PSTPIP1 rescue cell lines, we found that there was no significant difference in PTP-PEST interaction between PSTPIP1-WT and -R405C (Figure 6D-E). As a confirmation of this finding, we tested the phosphorylation of PSTPIP1, which is elevated if its interaction with PTP-PEST is disrupted.48 The PSTPIP1-R405C mutant was phosphorylated at a nearly identical level to that of PSTPIP1-WT, further supporting that this mutation does not alter PTP-PEST binding (Figure 6F-G). Because PSTPIP1 also binds to ABL, which is a regulator of podosomes and filopodia, we tested ABL phosphorylation in the PSTPIP1-WT and -R405C rescue cells and did not observe any differences (supplemental Figure 5B-C).51-53
Elevated WASP activity decreases podosomes and increases filopodia
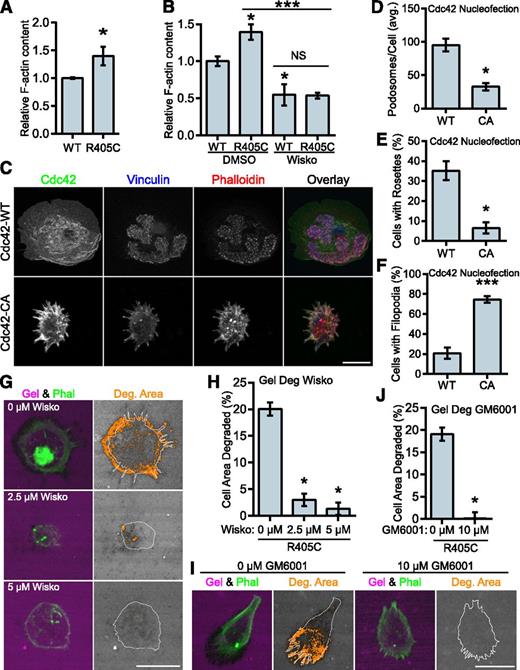
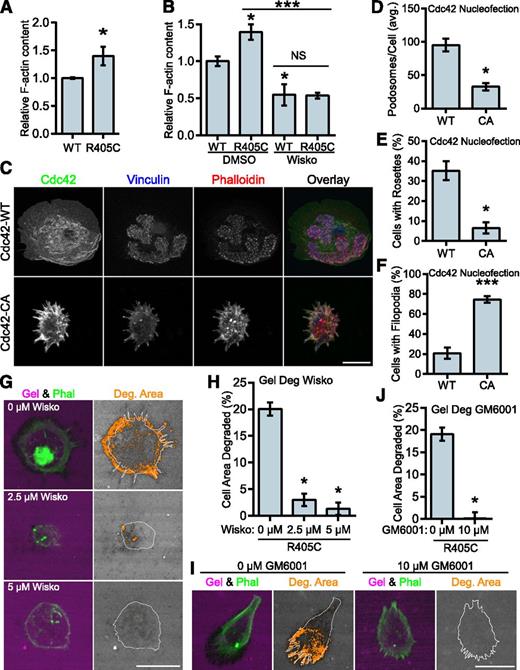
PSTPIP1 has been positioned as the link between PTP-PEST and WASP, which would allow it to indirectly modulate WASP activity.48 To determine whether the PSTPIP1-R405C mutant affected the regulation of WASP activity, we performed F-actin content assays. The R405C rescue cells had greater F-actin content than their WT counterparts (Figure 7A). Wiskostatin is a well-characterized inhibitor of WASP and N-WASP that stabilizes the autoinhibited conformation of these proteins, although some nonspecific effects have been reported.54-57 We found that wiskostatin treatment decreased the amount of F-actin in the cells. The amount of F-actin was the same in wiskostatin-treated WT and R405C rescue cells, indicating that the difference in F-actin was likely due to a difference in WASP or N-WASP activity (Figure 7B). Expression of constitutively active Cdc42 (Cdc42-CA) can be used to drive WASP activity.54 Although Cdc42 can activate WASP and N-WASP, it is a much stronger activator of WASP.58 It was previously demonstrated, but not rigorously quantified, that expression of Cdc42-CA could promote podosome disassembly and the formation of filopodia.17,59 We confirmed these findings by nucleofecting Cdc42-WT and -CA into primary human macrophages. We found that Cdc42-CA significantly decreased podosome formation with a concomitant increase in filopodia formation (Figure 7C-F).
WASP mediates the transition to filopodia in PSTPIP1-R405C cells. (A) The F-actin polymerizing ability of PSTPIP1-WT and -R405C rescue cells was determined by stimulating serum-starved cells with chemokine (C-C motif) ligand 2. F-actin was saturated with rhodamine phalloidin and extracted with methanol; fluorescence was determined with a plate reader. Fluorescence intensity was normalized to the level of PSTPIP1-WT cells for each experiment. (B) F-actin content assay performed as in (A). Prior to stimulation, cells were pretreated for 30 minutes with dimethyl sulfoxide (DMSO) or 10 μM wiskostatin. Stimulation was performed in the presence of DMSO or 20 μM wiskostatin (Wisko) for 5 minutes. DMSO-treated WT was the reference value for statistical comparison except where indicated by a bar. (C-F) Macrophages nucleofected with GFP-Cdc42 WT (WT) or GFP-Cdc42 Q61L (CA) were plated on fibrinogen-coated coverslips overnight and stained with rhodamine phalloidin, anti-vinculin antibody, and anti-GFP antibody to visualize podosomes and Cdc42 expression. (C) Representative images of podosomes and filopodia in Cdc42-nucleofected cells. GFP-positive cells were used for (D) quantification of the average number of podosomes per cell, (E) the percentage of cells that made podosome rosettes, and (F) the percentage of cells forming filopodia. (G-J) THP-1 shPST + R405C rescue cells were plated on Alexa Fluor 568–conjugated gelatin for 3 hours with the indicated concentration of (G) wiskostatin or (I) GM6001. Representative images are shown. (H) The amount of degradation in the presence of wiskostatin is shown as a percentage of cell area (0 μM, n = 112; 2.5 μM, n = 134; 5 μM, n = 138). (J) The amount of degradation in the presence of GM6001 is shown as a percentage of cell area (0 μM, n = 125; 10 μM, n = 131). All values are mean ± SEM from 3 (B,H,J) or 4 (A,D-F) independent experiments. *P < .05; ***P < .001 as determined by (A) unpaired Student t test, (B) one-way ANOVA with Sidak’s multiple comparison test, (D-F) paired Student t test, or (H,J) ANOVA with repeated measures using a compound symmetry correlation structure.
WASP mediates the transition to filopodia in PSTPIP1-R405C cells. (A) The F-actin polymerizing ability of PSTPIP1-WT and -R405C rescue cells was determined by stimulating serum-starved cells with chemokine (C-C motif) ligand 2. F-actin was saturated with rhodamine phalloidin and extracted with methanol; fluorescence was determined with a plate reader. Fluorescence intensity was normalized to the level of PSTPIP1-WT cells for each experiment. (B) F-actin content assay performed as in (A). Prior to stimulation, cells were pretreated for 30 minutes with dimethyl sulfoxide (DMSO) or 10 μM wiskostatin. Stimulation was performed in the presence of DMSO or 20 μM wiskostatin (Wisko) for 5 minutes. DMSO-treated WT was the reference value for statistical comparison except where indicated by a bar. (C-F) Macrophages nucleofected with GFP-Cdc42 WT (WT) or GFP-Cdc42 Q61L (CA) were plated on fibrinogen-coated coverslips overnight and stained with rhodamine phalloidin, anti-vinculin antibody, and anti-GFP antibody to visualize podosomes and Cdc42 expression. (C) Representative images of podosomes and filopodia in Cdc42-nucleofected cells. GFP-positive cells were used for (D) quantification of the average number of podosomes per cell, (E) the percentage of cells that made podosome rosettes, and (F) the percentage of cells forming filopodia. (G-J) THP-1 shPST + R405C rescue cells were plated on Alexa Fluor 568–conjugated gelatin for 3 hours with the indicated concentration of (G) wiskostatin or (I) GM6001. Representative images are shown. (H) The amount of degradation in the presence of wiskostatin is shown as a percentage of cell area (0 μM, n = 112; 2.5 μM, n = 134; 5 μM, n = 138). (J) The amount of degradation in the presence of GM6001 is shown as a percentage of cell area (0 μM, n = 125; 10 μM, n = 131). All values are mean ± SEM from 3 (B,H,J) or 4 (A,D-F) independent experiments. *P < .05; ***P < .001 as determined by (A) unpaired Student t test, (B) one-way ANOVA with Sidak’s multiple comparison test, (D-F) paired Student t test, or (H,J) ANOVA with repeated measures using a compound symmetry correlation structure.
We also predicted that if the phenotype observed in the R405C rescue cells was due to elevated WASP-family activity, then it could be reversed by WASP inhibition. We tested this by performing gelatin degradation assays in the presence of wiskostatin. We found that wiskostatin reduced gelatin degradation in the R405C cells in a dose-dependent manner. Wiskostatin treatment also reduced the formation of filopodia, indicating their dependence on WASP-family activity (Figure 7G-H). Because podosome-mediated gelatin degradation is matrix metalloprotease (MMP) dependent,14 we sought to determine whether the elevated gelatin degradation observed in the R405C cells was MMP dependent. Treatment with GM6001, a broad spectrum MMP inhibitor, decreased gelatin degradation in the R405C rescues without affecting filopodia, indicating that these cells degrade ECM through MMP activity (Figure 7I-J). Our observations indicate that despite the decrease in podosomes formed by the R405C rescue cells, the same pathways are used by filopodia-rich R405C rescue cells to degrade the ECM.
Discussion
Our results show that PSTPIP1 is a key adaptor protein that modulates the transition from podosomes to filopodia-like protrusions and regulates invasive macrophage migration. Our findings also highlight how the study of a patient with a rare autoinflammatory disease can provide insight into how cell motility and the cytoskeleton are regulated and altered in disease. Finally, this study raises the intriguing idea that matrix degradation and tissue damage by highly degradative macrophages may be important in the pathogenesis of pyoderma gangrenosum and raises interesting questions about new targets for treatment.
The PAPA macrophages used in this study transitioned from formation of podosomes to filopodia-like protrusions with an associated increase in degradation and invasive migration. This is in contrast to our previous report on PAPA patients with the A230T mutation, in which macrophages had fewer podosomes, impaired matrix degradation, and decreased invasion.29 Our data suggested that the filopodia induced by PSTPIP1-R405C are capable of degrading ECM as a result of the presence of filopodia over degraded areas. It is interesting to note that the PSTPIP1-R405C cells can also make prominent vinculin-containing focal contacts that may also contribute to localized matrix degradation. Our observation that macrophages often exclusively form podosomes or filopodia-like projections suggests that there is a cellular switch controlling the transition between these structures.
WASP, due to its key role in regulating F-actin polymerization and its necessity for both podosomes and filopodia,60 is well positioned to be the switch that can fine-tune these actin-based structures. WASP is necessary for podosome formation in macrophages and dendritic cells, and mutations that affect its expression, stability, or activity severely impair podosome formation.17,24 Several approaches have demonstrated that WASP can regulate the organization of podosomes in addition to being necessary for their formation. Podosome formation, organization, and degradative ability can be enhanced by increasing WASP phosphorylation or introducing a phospho-mimic version of WASP into macrophages.13,61 Additionally, PTP-PEST–deficient, src-transformed fibroblasts have increased podosome rosette formation.62
WASP activity can also be modulated to induce filopodia formation. In addition to promoting podosome formation, WASP-Y291E (phospho-mimic) also drives filopodia formation in macrophages.63 Additionally, macrophages injected with constitutively active Cdc42 produced fewer podosomes and more filopodia than controls.17,59 In this study, we demonstrated that ectopic expression of Cdc42-CA produced a decrease in podosomes and an increase in filopodia compared with expression of Cdc42-WT. However, we did not observe an increase in matrix degradation with ectopic expression of Cdc42-CA alone (data not shown), suggesting that there are other PSTPIP1-regulated proteins that may play a role in this transition. Together these data suggest that there is a continuum of WASP activity in macrophages that correlates with the formation of different actin-based structures (supplemental Figure 6).
Our data suggest that the PSTPIP1-R405C mutant permits excessive activation of WASP, causing a transition from podosome to filopodia formation. PSTPIP1-R405C had significantly impaired interaction with WASP, but its interaction with PTP-PEST was unaltered. Additionally, the increased F-actin content, gelatin degradation, and filopodia formation in the R405C cells were reversed by WASP inhibition. PSTPIP1 bridges the interaction of WASP and PTP-PEST,48 and our data suggest that PSTPIP1-R405C can no longer perform this function, leading to elevated WASP activity. Unfortunately, we were unable to detect differences in WASP phosphorylation in THP-1 cells expressing PSTPIP1-WT and -R405C, despite taking several approaches. Because PSTPIP1-R405C can still bind to PTP-PEST, it may sequester this WASP-inhibitory factor away from WASP, causing an additional increase in WASP activity and leading to filopodia formation (supplemental Figure 6). Moreover, the presence of PSTPIP1 on the dorsal surface opposite of podosomes, suggests that PSTPIP1 may normally sequester WASP away from podosomes to limit its activity. The PSTPIP1-R405C mutation may also allow WASP to relocate in a manner that promotes filopodia formation.
Our results also indicate that PSTPIP1 can interact with N-WASP, and further experiments will be necessary to determine whether the interaction is altered by the PSTPIP1-R405C mutation. WASP and N-WASP perform nonredundant roles in macrophage podosomes because WASP controls podosome formation and organization, while N-WASP regulates ECM degradation independent of podosome formation.64 These findings suggest that the differences in podosome organization we observed are most likely attributable to WASP rather than N-WASP.
Together, our data and the published literature support the idea that WASP plays a critical role in regulating the transition from individual podosomes to podosome rosettes to filopodium formation. We believe that the transition from podosomes to filopodia may be important for invasive migration through tissues and may contribute to pathologic degradation. This idea is supported by studies of N-WASP in cell migration. N-WASP is dispensable for 2D migration on a rigid substrate; however, N-WASP–deficient breast cancer cells cannot form long, invasive protrusions or invade through 3D matrix.65 WASP-deficient macrophages and dendritic cells have reduced 2D chemotaxis but maintain their ability to randomly migrate.13,18,19 To the best of our knowledge, the role of WASP in 3D-invasive migration has not been directly tested. However, hematopoietic cell kinase, which activates WASP, localizes to invasive 3D podosomes.25 We found that the PAPA patient macrophages were more degradative and more invasive and that they produce filopodia-like structures similar to the previously described 3D podosomes. We believe that the role of PSTPIP1 in 3D podosome function would be an interesting area for future study. Thus, our data suggest that elevated WASP activity drives macrophages to adopt a morphology characterized by filopodia formation and that PSTPIP1 regulates this transition.
In this study, we have characterized the role of PSTPIP1 and the disease-associated R405C mutation on actin-based protrusive structures. We believe that our observations demonstrating increased matrix degradation and formation of filopodia-like structures in PAPA patient macrophages have important clinical implications. Pyoderma gangrenosum, the severe ulcerative lesion seen in PAPA syndrome, is considered a neutrophilic dermatosis.7 However, our findings that PAPA macrophages are highly invasive and degradative suggest that the tissue damage they cause could lead to subsequent neutrophil infiltration. Pyoderma gangrenosum is frequently associated with inflammatory bowel disease, but pyoderma gangrenosum lesions can be present even if the underlying disease is in remission and other inflammatory parameters are low.7,66,67 Accordingly, the pyoderma gangrenosum in our patient did not improve even after treatment with immunosuppressive medications had returned inflammatory parameters to normal levels. These observations suggest that systemic inflammation and tissue damage in pyoderma gangrenosum are separable, and that the most successful treatment modalities would address both problems. We found that the elevated matrix degradation seen in the R405C rescue cells could be reversed by WASP or MMP inhibition, which could be rational targets for preventing tissue damage in pyoderma gangrenosum. Finally, our findings indicate the importance of PSTPIP1 in regulating WASP activity in the context of a severe autoinflammatory disease. While decreased WASP activity has long been associated with immunodeficiency, our findings associate elevated WASP activity with autoinflammatory disease. We have shown that disruption of the PSTPIP1-WASP interaction increases WASP activity, leading to increased formation of filopodia with a corresponding increase in invasion and degradation. Overall, our findings suggest that the transition to a highly invasive phenotype under control of PSTPIP1 may be an important factor in the pathogenesis of PAPA syndrome.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors would like to thank the National Heart, Lung, and Blood Institute Grand Opportunity (GO) Exome Sequencing Project and its ongoing studies which produced and provided exome variant calls for comparison: the Lung GO Sequencing Project (HL-102923), the Women's Health Initiative Sequencing Project (HL-102924), the Broad GO Sequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926) and the Heart GO Sequencing Project (HL-103010). Thanks to Sa Kan Yoo and Danny LeBert for critically reading this manuscript.
This work was supported by National Institutes of Health (NIH), National Cancer Institute grant CA085862 (A.H.), a grant from the Burroughs Wellcome fund (A.H.), and NIH grant AR059703 (P.J.F.). TWS received predoctoral funding support from an NIH, National Cancer Institute F30 fellowship (NIH grant HL114143), the University of Wisconsin Hematology training grant (NIH grant HL007899), and the University of Wisconsin Medical Scientist Training Program training grant (NIH grant GM008692).
Authorship
Contribution: T.W.S. and A.H. designed the studies, analyzed data, and wrote the manuscript; T.W.S., D.A.B., X.B., D.C.G., and P.J.F. performed experiments and analyzed data; J.C.E. performed statistical analysis; and J.M.B. and C.M.S. contributed patient data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Anna Huttenlocher, Department of Medical Microbiology and Immunology, 1550 Linden Dr, Room 4205 Microbial Sciences Building, Madison, WI 53706; e-mail: huttenlocher@wisc.edu.