Key Points
miR-146a and miR-146b are upregulated during premalignancy in the thymus of T cell–specific PTEN-deficient mice.
Transgenic expression of mir-146a/b delays PTEN-deficient lymphomagenesis through repression of TCR signals critical for c-myc activation.
Abstract
Cancer develops by a multistep process during which cells acquire characteristics that allow them to evade apoptosis and proliferate unchecked. Sequential acquisition of genetic alterations drives this process but also causes cellular stress, frequently prompting cells to enter a premalignant period during which they mount a defense against transformation. T cell–specific deletion of the tumor suppressor PTEN in mice induces premalignancy in the thymus and development of CD4+ T-cell lymphomas in the periphery. Here we sought to identify factors mediating the cellular defense against transformation during the premalignant period. We identified several microRNAs upregulated specifically in premalignant thymocytes, including miR-146a, miR-146b, and the miR-183/96/182 cluster. CD4-driven T cell–specific transgenic overexpression of mir-146a and mir-146b significantly delayed PTEN-deficient lymphomagenesis and delayed c-myc oncogene induction, a key driver of transformation in PTEN-deficient T-cell malignancies. We found that miR-146a and miR-146b targeting of Traf6 attenuates TCR signaling in the thymus and inhibits downstream NF-κB–dependent induction of c-myc. Additionally, c-myc repression in mature CD4 T cells by miR-146b impaired TCR-mediated proliferation. Hence, we have identified 2 miRNAs that are upregulated as part of the cellular response against transformation that, when overrepresented, can effectively inhibit progression to malignancy in the context of PTEN deficiency.
Introduction
Malignant cancer is typically preceded by the development of premalignant lesions. However, not all premalignant lesions progress to malignancy.1 Premalignant cells arise in large part because of genetic alterations that promote excessive growth and proliferation.2 Characterizing changes in gene expression that occur in premalignant lesions could aid in correlating cellular responses to transformation with the risk of disease progression.2 Furthermore, it could lead to identification of factors mediating the antitumor response that might inspire the design of more effective targeted therapeutics. Although the biological heterogeneity of human premalignant lesions makes them difficult to study, premalignant lesions have been identified in many genetic mouse models of human cancer. The genetic homogeneity of mice can lead to more synchronous development of homogenous lesions, greatly facilitating the study of premalignancy.3
Phosphatase and tensin homolog (PTEN) is one of the most frequently inactivated tumor suppressors in human cancer.4 PTEN dephosphorylates 3-phosphoinositide products of PI3 kinase (PI3K), thereby negatively regulating PI3K-Akt signaling, which promotes cell growth and proliferation.5,6 Pten is inactivated in many cancers by loss of heterozygosity, mutation, or deletion, with a high incidence in glioma, breast cancer, melanoma, prostate cancer, leukemia, and lymphoma.4 In T-cell acute lymphoblastic leukemia/lymphoma (T-ALL), Pten mutations have been identified in as much as 27% of patients analyzed.7 Deletions are found in >8% of patients and have been associated with early treatment failure.7,8 Consequential hyperactivation of the PI3K-Akt pathway is also commonly observed.9 For cases in which Pten is not altered at the genomic level, PTEN expression is still frequently downregulated.8
T cell–specific deletion of PTEN in mice induces premalignancy in CD4+CD8+ (double-positive [DP]) thymocytes, which progresses to CD4+ T-cell lymphoma in the lymph nodes and spleen.10-12 Interestingly, premalignancy occurs only in animals 6 weeks or older. In younger mice, T-cell development in T cell–specific PTEN-deficient mice (tPTEN−/−) is completely normal, with no signs of malignancy.10 Premalignant thymocytes harbor T-cell receptor (TCR) signaling–dependent TCRα/c-myc chromosomal translocations that promote c-myc upregulation, critical for promoting transformation of these cells.11 Premalignancy is also characterized by increased activation of the PI3K-Akt signaling pathway and induction of a senescence program.10 However, because DP thymocytes do not proliferate to any significant degree, it is unlikely that senescence acts as a barrier to transformation in this model.10 Thus we sought to identify other factors that stave off transformation in premalignant PTEN-deficient DP cells.
Through microRNA (miRNA) expression analyses, we have identified miRNAs miR-146a and miR-146b as being significantly upregulated in premalignant DP cells of tPTEN−/− mice. Strikingly, T cell–specific expression of mir-146a and mir-146b transgenes significantly delayed tPTEN−/− lymphomagenesis, supporting their expression as a barrier to transformation. Tumor suppression was mediated by miR-146 attenuation of TCR signaling through repression of its target Traf6, an important activator of NF-κB. Our results not only support the potential therapeutic applications of these tumor-suppressive miRNAs but also suggest a general strategy for the identification of miRNAs that inhibit transformation in other cancer models.
Materials and methods
Mice
Characterization of the Lck-Cre Ptenfl/fl (tPTEN−/−) mice has previously been reported.10,13 Analyses were performed on a mixed 129/SvJ × CBA × C57BL/6 background or a C57BL/6 background (backcrossed >10 generations). All of the animal-related procedures have been approved by the UC Berkeley Animal Care and Use Committee.
The miRNA transgenic mouse constructs were generated by cloning polymerase chain reaction (PCR)-amplified pre-miRNA sequences, with approximately 450bp of flanking sequence into the pTG4 construct.14,15 The primer sequences used were: miR-146a-forward ATCGATCGCTCGAGTCTGGCTAGCCTGG, miR-146a-reverse ATCGATCGCTCGAAATAGGCGTTGAG, miR-146b-forward ATCGATCGCTCGAGCCATAGGCTGTGAT, miR-146b-reverse ATCGATCGCTCGACCCACAGGAGGAT. Founders were identified by PCR genotyping with miR-146a-forward, miR-146b-forward, and miR-146a/b-reverse TGTGCCAAAGGGATTTTAGG.
miRNA microarray
DP thymocytes were collected by cell sorting. Total RNA was extracted with a miRNeasy Mini Kit (Qiagen, Valencia, CA). RNA was submitted to Exiqon (Woburn, MA) for quality control, labeling with miRCURY Hy3/Hy5 probes, and hybridization onto a miRCURY LNA Array (v.11.0 mmu). Data analysis was performed by Exiqon (http://mcb.berkeley.edu/labs/winoto/array.html). (Accession number for the miRNA array data at the GEO public database is GSE56759.)
Quantitative real-time (RT) PCR
DP and CD4 SP thymocytes were collected by cell sorting, and mature T cells were collected by negative selection column purification or cell sorting. Taqman MicroRNA Assays (Applied Biosystems, Life Technologies, Grand Island, NY) specific to each miRNA were used for miRNA quantitation. For mRNA quantitation, reverse transcription was performed using SuperScript III with Oligo(dT) primers (Invitrogen, Life Technologies). SYBR Green (Invitrogen) RT-PCR was performed using the following primers: GAPDH-forward ATCTTCTTGTGCAGTGCCAGCCTCGTCCCG, GAPDH-reverse AGTTGAGGTCAATGAAGGGGTCGTTGATGG, c-myc-forward GCCCCTAGTGCTGCATGA, c-myc-reverse AGAAGATGAGGAAGAAATTGATGTGG, Traf6-forward TTTCCCTGACGGTAAAGTGC, and Traf6-reverse CTGGCACTTCTGGAAAGGAC.
RNase protection assay
miR-146a and miR-16 probes were synthesized and 32P-labeled using the miRVana miRNA Probe Construction Kit (Ambion, Life Technologies) with the miR-16 DNA oligonucleotides provided in the kit and the following miR-146a DNA oligonucleotides as templates: sense TGAGAACTGAATTCCATGGGTTCCTGTCTC, anti-sense GAAGAACTGAATTTCACAGGTCCCTGTCTC. The mirVana miRNA Detection Kit was used for probe hybridization and RNase A/RNase T1 digestion. Protected fragments were run on a polyacrylamide gel and visualized by radiographic film. Yeast RNA was used as a negative control.
Results
Upregulation of several miRNAs correlates with premalignancy in PTEN-deficient DP thymocytes
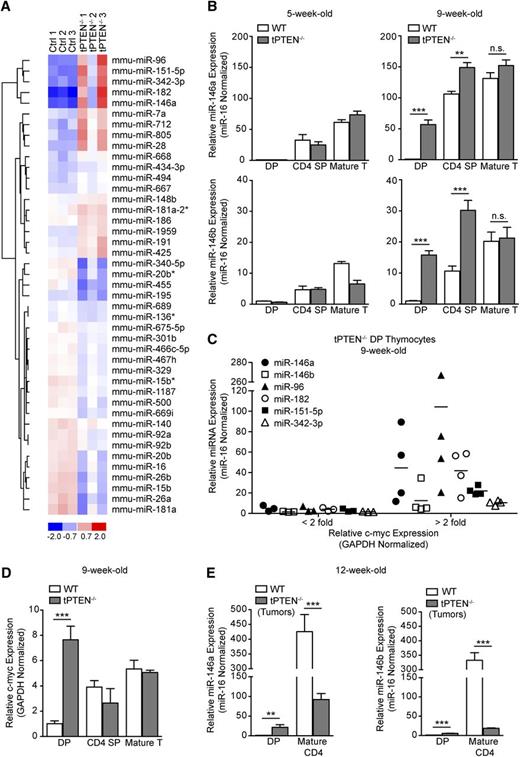
To identify miRNAs that might act to inhibit transformation of PTEN-deficient DP thymocytes, we performed a miRNA microarray on sorted DP cells from three 9-week-old tPTEN−/− mice and 3 littermate controls (Figure 1A). Nine-week-old tumor-free tPTEN−/− mice were selected based on our prior correlation of premalignancy with this age.10 We observed that 43 of 599 miRNAs were differentially expressed in the tPTEN−/− samples. Quantitative RT-PCR validation of these results indicated that several miRNAs were upregulated specifically in 9-week-old tPTEN−/− DP and sometimes CD4 SP thymocytes, but not in the corresponding mature T cells (Figure 1B and supplemental Figure 1A). Interestingly, upregulation was not detected in DP thymocytes from 5-week-old mice, where premalignancy has not yet occurred, indicating that these miRNAs are specifically induced during the premalignant period. We identified miR-146a as one of the most highly upregulated miRNAs in 9-week-old tPTEN−/− DP thymocytes. Expression of highly homologous family member miR-146b was also increased. Although encoded on different chromosomes, miR-146a and miR-146b have identical seed sequences and differ by only 2 nucleotides at the 3′ end and are therefore predicted to have similar mRNA targets. Also highly upregulated in 9-week-old DP thymocytes were the miRNAs of the miR-183/96/182 cluster, miR-151-5p and miR-342-3p (supplemental Figure 1A). We hypothesized that one or more of these miRNAs might be upregulated to inhibit malignant transformation of PTEN-deficient DP thymocytes.
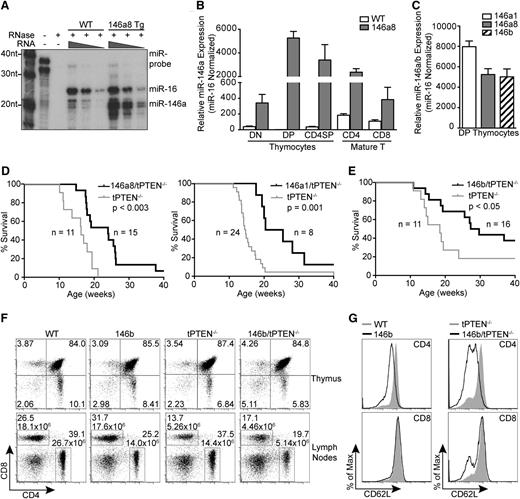
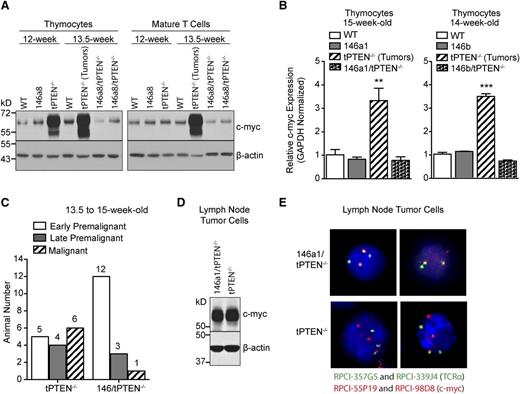
Several miRNAs are highly upregulated in premalignant tPTEN−/− DP thymocytes. (A) Heat-map representing miRNAs with a combined P value of < .05 and a δ log median ratio >0.2. Forty-three of 599 miRNAs evaluated were differentially expressed in tPTEN−/− DP thymocytes from 3 premalignant 9-week-old mice compared with 3 littermate controls (129/SvJ × CBA × C57BL/6 background). Red and blue indicate miRNA expression levels above and below the mean, respectively. (B) Quantitative RT-PCR validation of the miRNA array results on fluorescence-activated cell sorting (FACS)-sorted DP and CD4 SP thymocytes and column-purified mature T cells from 5-week-old and premalignant 9-week-old mice. miRNA expression levels are normalized to miR-16, which is expressed similarly in WT and tPTEN−/− T-cell subsets (supplemental Figure 1B). Fold change is relative to WT DP thymocytes. (C) Quantitative RT-PCR analysis of the correlation between miRNA and c-myc expression levels in 9-week-old tPTEN−/− premalignant DP thymocytes. Fold change is relative to littermate WT DP thymocytes. c-myc expression is represented as less than or greater than a twofold increase over WT levels. (D) Quantitative RT-PCR analysis of c-myc expression in DP and CD4 SP thymocytes and mature T cells from premalignant 9-week-old mice with a pattern of miR-146a and miR-146b expression as shown in (B). (E) miR-146a and miR-146b expression levels in thymocytes and lymph node tumor cells from 12-week-old mice (129/SvJ × CBA × C57BL/6 background). Fold change is relative to WT DP thymocytes. Data in (B) and (D-E) are presented as mean ± SD and are representative of 3 or more independent experiments. Statistics were calculated by Student t test (***P < .001, **P < .01, *P < .05, n.s. not significant).
Several miRNAs are highly upregulated in premalignant tPTEN−/− DP thymocytes. (A) Heat-map representing miRNAs with a combined P value of < .05 and a δ log median ratio >0.2. Forty-three of 599 miRNAs evaluated were differentially expressed in tPTEN−/− DP thymocytes from 3 premalignant 9-week-old mice compared with 3 littermate controls (129/SvJ × CBA × C57BL/6 background). Red and blue indicate miRNA expression levels above and below the mean, respectively. (B) Quantitative RT-PCR validation of the miRNA array results on fluorescence-activated cell sorting (FACS)-sorted DP and CD4 SP thymocytes and column-purified mature T cells from 5-week-old and premalignant 9-week-old mice. miRNA expression levels are normalized to miR-16, which is expressed similarly in WT and tPTEN−/− T-cell subsets (supplemental Figure 1B). Fold change is relative to WT DP thymocytes. (C) Quantitative RT-PCR analysis of the correlation between miRNA and c-myc expression levels in 9-week-old tPTEN−/− premalignant DP thymocytes. Fold change is relative to littermate WT DP thymocytes. c-myc expression is represented as less than or greater than a twofold increase over WT levels. (D) Quantitative RT-PCR analysis of c-myc expression in DP and CD4 SP thymocytes and mature T cells from premalignant 9-week-old mice with a pattern of miR-146a and miR-146b expression as shown in (B). (E) miR-146a and miR-146b expression levels in thymocytes and lymph node tumor cells from 12-week-old mice (129/SvJ × CBA × C57BL/6 background). Fold change is relative to WT DP thymocytes. Data in (B) and (D-E) are presented as mean ± SD and are representative of 3 or more independent experiments. Statistics were calculated by Student t test (***P < .001, **P < .01, *P < .05, n.s. not significant).
Analysis of a larger cohort of 9-week-old tPTEN−/− mice, however, revealed that these miRNAs are not always as highly upregulated at this age, consistent with the more modest phenotype observed for sample tPTEN−/− 2 in the miRNA microarray. Analysis of c-myc expression in these mice uncovered a striking correlation between miRNA upregulation and high c-myc expression (Figure 1C).11 c-myc upregulation was only observed in DP thymocytes and not in more mature thymocytes or T cells, paralleling the miRNA expression pattern we observed (Figure 1D). However, it is unlikely that c-myc drives expression of these miRNAs because prior analysis of c-myc regulation of global miRNA expression by others found that c-myc activation does not increase their expression.16,17
In 12-week-old tPTEN−/− mice harboring CD4+ T-cell lymphomas, miR-146a and -146b expression in sorted CD4+ mature T cells was often downregulated (Figure 1E). This expression pattern is consistent with that observed for tumor-suppressive miRNAs, such as miR-34a or let-7, because their downregulation is advantageous to the tumor.18 In contrast, the miR-183/96/182 cluster was further upregulated in tumor cells (data not shown).
Generation of T cell–specific miRNA transgenic mice
To test whether upregulation of miR-146a or miR-146b might act to inhibit malignant transformation of tPTEN−/− DP thymocytes, we generated T cell–specific mir-146a and mir-146b transgenic mice. The miRNA transgenes were expressed under the control of the CD4 regulatory elements as previously described.14,15 The transgenic mice were generated on the C57BL/6 background and 2 146a founder lines (a1 and a8) and 1 146b founder line were used for analysis.
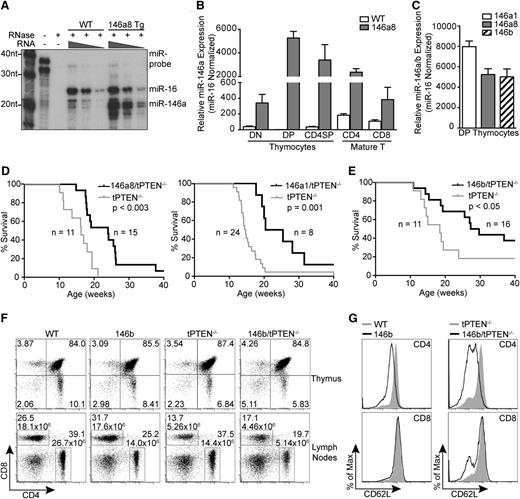
By RNase protection assay, we observed that miR-146a is normally expressed at low levels in thymocytes. In the mir-146a transgenic mice, miR-146a expression is increased to a level similar to that of miR-16, a miRNA that is abundant in thymocytes (Figure 2A).19 This increase in miR-146a expression is lymphoid tissue–specific because no upregulation was observed in the liver or kidneys (supplemental Figure 2A). The highest levels of mir-146a transgene expression were detected in DP thymocytes, followed by CD4 SP thymocytes and CD4+ mature T cells (Figure 2B). Some mir-146a transgene expression was also observed in CD4–CD8– (double-negative [DN]) thymocytes and CD8+ mature T cells. Importantly, the level of miR-146 transgene expression in DP thymocytes was similar between all of the transgenic lines analyzed (Figure 2C).
Transgenic expression of mir-146a and mir-146b delays tumorigenesis in tPTEN−/− mice. (A) miR-146a expression levels in total thymocytes from mir-146a transgenic line 8 (146a8) vs WT mice by RNase protection assay. miR-16, a miRNA that is abundant in thymocytes, was used as a reference for miRNA levels. (B) Quantitative RT-PCR analysis of miR-146a expression levels in FACS-sorted thymocyte and mature T-cell subsets from 146a8 transgenic and littermate WT control mice. Fold change is relative to WT DP thymocytes and normalized to miR-16. (C) Comparison of miR-146 expression levels in DP thymocytes from all characterized mir-146 transgenic founder lines. Fold change is relative to miR-146 expression in littermate WT controls. (D-E) Kaplan-Meier curves for the mir-146a transgenic lines 146a8 and 146a1 (D) and the mir-146b transgenic line (E) crossed to tPTEN−/− mice on the C57BL/6 background. Statistical significance was calculated using the Gehan-Breslow-Wilcoxon method. (F) CD4 vs CD8 flow cytometric analysis of T-cell populations in the thymus and lymph nodes of 7-week-old 146b transgenic mice compared with littermate controls on both WT and tPTEN−/− backgrounds. T-cell numbers are indicated on the lymph node plots to reflect the decrease in CD4 T-cell numbers with mir-146b transgene expression. Data are representative of 3 independent experiments. (G) Flow cytometric analysis of CD62L (l-selectin) surface expression in lymph node T cells from 7-week-old 146b transgenic and littermate control mice on WT and tPTEN−/− backgrounds. Data are representative of 2 independent experiments.
Transgenic expression of mir-146a and mir-146b delays tumorigenesis in tPTEN−/− mice. (A) miR-146a expression levels in total thymocytes from mir-146a transgenic line 8 (146a8) vs WT mice by RNase protection assay. miR-16, a miRNA that is abundant in thymocytes, was used as a reference for miRNA levels. (B) Quantitative RT-PCR analysis of miR-146a expression levels in FACS-sorted thymocyte and mature T-cell subsets from 146a8 transgenic and littermate WT control mice. Fold change is relative to WT DP thymocytes and normalized to miR-16. (C) Comparison of miR-146 expression levels in DP thymocytes from all characterized mir-146 transgenic founder lines. Fold change is relative to miR-146 expression in littermate WT controls. (D-E) Kaplan-Meier curves for the mir-146a transgenic lines 146a8 and 146a1 (D) and the mir-146b transgenic line (E) crossed to tPTEN−/− mice on the C57BL/6 background. Statistical significance was calculated using the Gehan-Breslow-Wilcoxon method. (F) CD4 vs CD8 flow cytometric analysis of T-cell populations in the thymus and lymph nodes of 7-week-old 146b transgenic mice compared with littermate controls on both WT and tPTEN−/− backgrounds. T-cell numbers are indicated on the lymph node plots to reflect the decrease in CD4 T-cell numbers with mir-146b transgene expression. Data are representative of 3 independent experiments. (G) Flow cytometric analysis of CD62L (l-selectin) surface expression in lymph node T cells from 7-week-old 146b transgenic and littermate control mice on WT and tPTEN−/− backgrounds. Data are representative of 2 independent experiments.
mir-146a and mir-146b transgene expression inhibits tPTEN−/− lymphomagenesis
To determine whether miR-146a and miR-146b overexpression can inhibit PTEN-deficient lymphomagenesis, the mir-146a and mir-146b transgenic lines were crossed to the tPTEN−/− mice. In preparation for the cross, the tPTEN−/− mice were backcrossed to C57BL/6 to match the background of the miRNA transgenic mice. The change in background somewhat lengthened the lifespan of the tPTEN−/− mice, with most mice succumbing to lymphoma by 20 weeks instead of 15 weeks. Strikingly, miR-146a or miR-146b overexpression significantly prolonged the survival of the tPTEN−/− mice, with a median increase in survival of 8 to 10 weeks (Figure 2D-E).
To elucidate the mechanism by which miR-146a and miR-146b delay tumorigenesis, we first evaluated whether overexpression of the miRNAs alters T-cell development. T-cell development occurs normally in the mir-146a transgenic lines, with no significant differences in T-cell proportions in the thymus or lymph nodes on wild-type (WT) or tPTEN−/− backgrounds (supplemental Figure 2B). mir-146b transgenic mice had normal thymic cell proportions; however, CD4 T-cell proportions were consistently reduced, with a twofold decrease in CD4 T-cell number in the lymph nodes regardless of PTEN status (Figure 2F). The mir-146b transgenic CD4 T cells had a modest but consistent reduction in surface expression of CD62L, which is normally upregulated upon thymic egress to mediate homing to the lymph nodes and spleen (Figure 2G). Reduced T-cell homing to the lymph nodes might contribute to the lower 146b CD4 T-cell numbers. In contrast, 146a8 CD4 T cells expressed normal levels of CD62L (supplemental Figure 2C). 146b CD4 T-cell maturation was otherwise generally unaffected, with appropriate upregulation of Qa-2 and downregulation of HSA. CD62L levels in SP thymocytes were also largely normal (supplemental Figure 2B-C). Thus, T-cell development appears to be largely unaltered by mir-146a and mir-146b transgene expression, with the exception of a moderate reduction in CD4 T-cell numbers in the 146b transgenic mice.
miR-146 overexpression delays c-myc upregulation associated with premalignancy
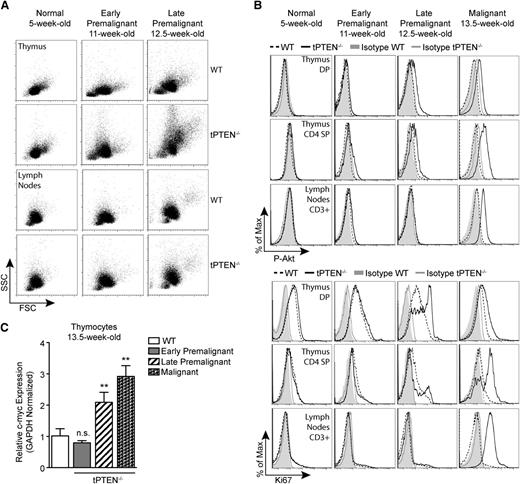
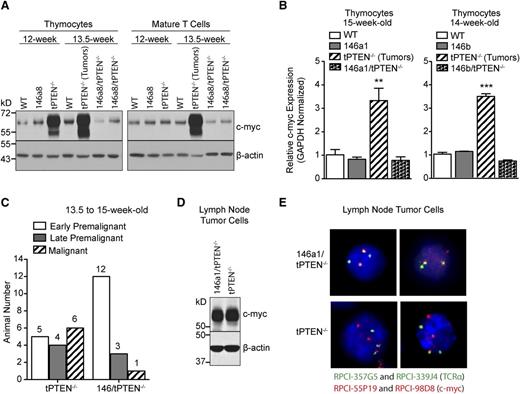
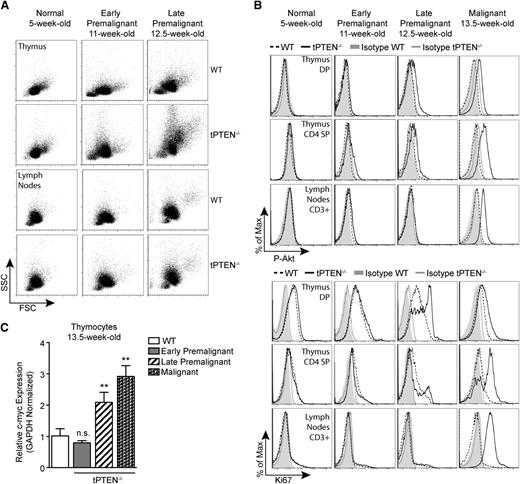
To evaluate whether miR-146 overexpression delays tPTEN−/− tumor formation, we first sought to characterize the premalignant state of tPTEN−/− mice on the C57BL/6 background. Consistent with the increased survival of C57BL/6 tPTEN−/− mice, the appearance of premalignant markers is delayed compared with tPTEN−/− mice on a mixed background.10 Beginning around 11 weeks, blasting, characterized by increased forward and side scatters, is apparent in the thymus, but not in the lymph nodes, of tPTEN−/− mice (early premalignant; Figure 3A). A slight increase in phosphorylated-Akt (P-Akt) is also evident in CD4 SP thymocytes (Figure 3A-B). Progression from early to late premalignancy is signified by more pronounced blasting and high levels of P-Akt in DP and CD4 SP thymocytes, but not in mature T cells (Figure 3B). Paralleling P-Akt levels, Ki67 expression is also increased in DP and CD4 SP thymocytes during late premalignancy (Figure 3B). Importantly, DP thymocytes normally express high levels of Ki67, even though they do not proliferate.10,14 Increased c-myc expression corresponds with late premalignancy, because increased c-myc mRNA levels are found only in thymocytes expressing high levels of both phosphorylated Akt and Ki67 (Figure 3C). Consistent with this observation, increased c-myc protein levels were observed in the thymus, but not in the lymph node T cells of 12-week-old late premalignant mice (Figure 4A). However, in 13.5-week-old tPTEN−/− mice that have developed tumors, c-myc upregulation was apparent in both compartments. Interestingly, no c-myc upregulation was observed in two 13.5-week-old littermate 146a8/tPTEN−/− mice in either compartment (Figure 4A). We also did not observe c-myc upregulation at the transcript level in thymocytes of 14- to 15-week-old tumor-free 146a1/tPTEN−/− and 146b/tPTEN−/− mice compared with littermate tPTEN−/− controls that had acquired tumors by this age (Figure 4B). These data suggest that miR-146 overexpression might act to delay c-myc upregulation associated with premalignancy. To more comprehensively assess the impact of miR-146 overexpression on premalignancy, we assessed blasting, P-Akt levels, Ki67 expression, and thymic c-myc mRNA expression in a large cohort of 13.5- to 15-week-old littermate or age-matched tPTEN−/− and 146/tPTEN−/− mice. These mice were then classified as early premalignant, late premalignant, or malignant based on the criteria outlined before. A large majority of the 146/tPTEN−/− mice were found to be in early premalignancy, compared with tPTEN−/− mice, which were primarily in late premalignancy or had already developed tumors (Figure 4C). These data suggest that miR-146 overexpression acts to prolong the survival of tPTEN−/− mice by delaying progression to malignancy or by inhibiting expansion of premalignant thymocytes. Consistent with increased c-myc mRNA levels in the thymus of late premalignant/malignant 146/tPTEN−/− mice, upregulation of c-myc expression and TCRα/c-myc translocations were found in 146/tPTEN−/− tumor cells (Figures 4D-E). Thus, miR-146 overexpression can delay, but not abolish, the hallmark events of tPTEN−/− lymphomagenesis.
c-myc upregulation occurs in late premalignancy and coincides with increased Akt signaling and Ki67 expression. (A) Forward scatter (FSC) and side scatter (SSC) analysis depicting increased blasting during premalignancy in the thymus, but not the lymph nodes, of tPTEN−/− mice compared with littermate WT controls. Blasting is more pronounced in late premalignancy. (B) Flow cytometric analysis of Akt phosphorylation at Ser473 (P-Akt) and Ki67 expression in DP and CD4 SP thymocytes and CD3+ lymph node T cells in littermate tPTEN−/− and WT mice. Late premalignancy is defined by a marked increase in both P-Akt and Ki67 in DP and CD4 SP thymocytes, but low expression of both markers in lymph node T cells. (C) Quantitative RT-PCR analysis showing the correlation of c-myc upregulation with late premalignancy in total thymocytes from littermate 13.5-week-old tPTEN−/− mice. Early premalignant, late premalignant, and malignant populations were distinguished by the extent of blasting and Ki67 expression, as described in (A) and (B), and peripheral tumor formation. Fold change is relative to WT thymocytes. Data are presented as mean ± SD and are representative of 3 or more independent experiments. Statistics were calculated by Student t test (**P < .01, n.s., not significant).
c-myc upregulation occurs in late premalignancy and coincides with increased Akt signaling and Ki67 expression. (A) Forward scatter (FSC) and side scatter (SSC) analysis depicting increased blasting during premalignancy in the thymus, but not the lymph nodes, of tPTEN−/− mice compared with littermate WT controls. Blasting is more pronounced in late premalignancy. (B) Flow cytometric analysis of Akt phosphorylation at Ser473 (P-Akt) and Ki67 expression in DP and CD4 SP thymocytes and CD3+ lymph node T cells in littermate tPTEN−/− and WT mice. Late premalignancy is defined by a marked increase in both P-Akt and Ki67 in DP and CD4 SP thymocytes, but low expression of both markers in lymph node T cells. (C) Quantitative RT-PCR analysis showing the correlation of c-myc upregulation with late premalignancy in total thymocytes from littermate 13.5-week-old tPTEN−/− mice. Early premalignant, late premalignant, and malignant populations were distinguished by the extent of blasting and Ki67 expression, as described in (A) and (B), and peripheral tumor formation. Fold change is relative to WT thymocytes. Data are presented as mean ± SD and are representative of 3 or more independent experiments. Statistics were calculated by Student t test (**P < .01, n.s., not significant).
miR-146 overexpression delays c-myc upregulation associated with premalignancy. (A) Western blot analysis showing c-myc upregulation in total thymocytes from late premalignant 12-week-old tPTEN−/− mice and both thymocytes and mature T cells from malignant 13.5-week-old tPTEN−/− mice (C57BL/6 background). C-myc upregulation is not detected in the thymocytes or mature T cells of littermate 13.5-week-old 146a8/tPTEN−/− mice. Data are representative of 2 independent experiments. (B) Quantitative RT-PCR analysis of c-myc expression in total thymocytes of 14- and 15-week-old mice. c-myc is upregulated in the thymocytes of tPTEN−/− mice (with tumors), but not in thymocytes from tumor-free littermate 146a1/tPTEN−/− or 146b/tPTEN−/− mice. Data are presented as mean ± SD and are representative of 2 independent experiments per transgenic line. Statistics were calculated by Student t test (***P < .001, **P < .01). (C) Classification of littermate or age-matched 13.5- to 15-week-old tPTEN−/− and 146/tPTEN−/− mice based on the criteria outlined in Figure 3 (data from all transgenic lines were pooled). (D) Western blot analysis of c-myc protein expression in lymph node tumor cells of 12.5-week-old littermate tPTEN−/− and 146a1/tPTEN−/− mice. Data are representative of 3 independent experiments. (E) Fluorescence in-situ hybridization showing TCRα/c-myc chromosomal translocations in interphase-arrested lymph node tumor cells from the tPTEN−/− and 146a1/tPTEN−/− mice in (D). Images were captured at original magnification ×40.
miR-146 overexpression delays c-myc upregulation associated with premalignancy. (A) Western blot analysis showing c-myc upregulation in total thymocytes from late premalignant 12-week-old tPTEN−/− mice and both thymocytes and mature T cells from malignant 13.5-week-old tPTEN−/− mice (C57BL/6 background). C-myc upregulation is not detected in the thymocytes or mature T cells of littermate 13.5-week-old 146a8/tPTEN−/− mice. Data are representative of 2 independent experiments. (B) Quantitative RT-PCR analysis of c-myc expression in total thymocytes of 14- and 15-week-old mice. c-myc is upregulated in the thymocytes of tPTEN−/− mice (with tumors), but not in thymocytes from tumor-free littermate 146a1/tPTEN−/− or 146b/tPTEN−/− mice. Data are presented as mean ± SD and are representative of 2 independent experiments per transgenic line. Statistics were calculated by Student t test (***P < .001, **P < .01). (C) Classification of littermate or age-matched 13.5- to 15-week-old tPTEN−/− and 146/tPTEN−/− mice based on the criteria outlined in Figure 3 (data from all transgenic lines were pooled). (D) Western blot analysis of c-myc protein expression in lymph node tumor cells of 12.5-week-old littermate tPTEN−/− and 146a1/tPTEN−/− mice. Data are representative of 3 independent experiments. (E) Fluorescence in-situ hybridization showing TCRα/c-myc chromosomal translocations in interphase-arrested lymph node tumor cells from the tPTEN−/− and 146a1/tPTEN−/− mice in (D). Images were captured at original magnification ×40.
miR-146 negatively regulates TCR signaling in thymocytes
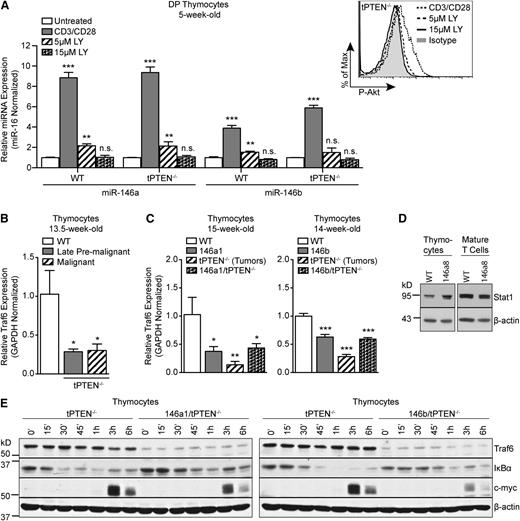
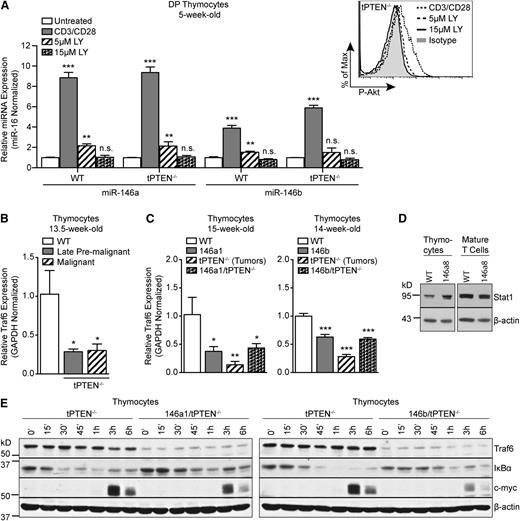
TCRα/c-myc translocations in PTEN-deficient thymocytes were previously demonstrated to be TCR signaling–dependent.11 In mature T cells, miR-146a expression is induced by TCR stimulation and miR-146a then acts in a negative feedback loop to repress TCR signaling.20,21 We hypothesized that miR-146a and miR-146b might similarly be induced by TCR signaling in DP thymocytes and act in a negative feedback loop to inhibit TCR signals necessary for TCRα/c-myc translocations/c-myc upregulation. Indeed, we found that miR-146a and miR-146b were significantly upregulated in TCR-stimulated DP thymocytes from both WT and tPTEN−/− 5-week-old mice (Figure 5A and supplemental Figure 3). However, miR-146b was upregulated less than miR-146a, consistent with less induction of miR-146b compared with miR-146a in premalignant tPTEN−/− DP thymocytes. Because strong amplification of Akt signaling is observed in late premalignant thymocytes, we tested whether Akt signaling downstream of the TCR might be responsible for miR-146a and miR-146b upregulation. Inhibition of Akt signaling with the PI3K inhibitor LY294002 almost completely abrogated miR-146a and miR-146b induction in TCR-stimulated DP thymocytes (Figure 5A and supplemental Figure 3). Thus Akt signaling downstream of the TCR is likely responsible for miR-146a and miR-146b upregulation during premalignancy in tPTEN−/− mice.
miR-146 represses TCR-dependent NF-κB signaling in thymocytes by targeting Traf6. (A) Quantitative RT-PCR analysis of miR-146a and miR-146b expression levels in FACS-sorted DP thymocytes, from 5-week-old littermate WT and tPTEN−/− mice (C57BL/6 background), left unstimulated or stimulated with anti-CD3/CD28 antibodies for 16 hours. Where indicated, DP thymocytes were pretreated with 5 μM or 15 μM LY294002 (LY) for 1 hour before anti-CD3/CD28 stimulation. Expression was normalized to miR-16 (or sno-234 in supplemental Figure 3) and fold change is relative to untreated cells. Flow cytometry analysis shows dose-dependent inhibition of Akt phosphorylation in LY294002-treated cells. (B) Quantitative RT-PCR analysis of Traf6 expression in littermate late premalignant and malignant tPTEN−/− mice compared with an age-matched WT control (C57BL/6 background). (C) Quantitative RT-PCR analysis of Traf6 expression in 146a1 and 146b transgenic mice compared with littermate controls on WT and tPTEN−/− backgrounds. (D) Western blot analysis of Stat1 protein expression in total thymocytes and mature T cells from 12-week-old littermate 146a8 transgenic and WT mice. (E) Western blot analysis of Traf6 expression, IκBα degradation, and c-myc induction in TCR-stimulated total thymocytes from 10-week-old 146a1/tPTEN−/− and 146b/tPTEN−/− mice and littermate or age-matched tPTEN−/− controls. Thymocytes were stimulated with PMA and ionomycin for the indicated times. Data in (A-C) are presented as mean ± SD and are representative of 3 independent experiments. Statistics were calculated by Student t test (***P < .001, **P < .01, *P < .05, n.s. not significant).
miR-146 represses TCR-dependent NF-κB signaling in thymocytes by targeting Traf6. (A) Quantitative RT-PCR analysis of miR-146a and miR-146b expression levels in FACS-sorted DP thymocytes, from 5-week-old littermate WT and tPTEN−/− mice (C57BL/6 background), left unstimulated or stimulated with anti-CD3/CD28 antibodies for 16 hours. Where indicated, DP thymocytes were pretreated with 5 μM or 15 μM LY294002 (LY) for 1 hour before anti-CD3/CD28 stimulation. Expression was normalized to miR-16 (or sno-234 in supplemental Figure 3) and fold change is relative to untreated cells. Flow cytometry analysis shows dose-dependent inhibition of Akt phosphorylation in LY294002-treated cells. (B) Quantitative RT-PCR analysis of Traf6 expression in littermate late premalignant and malignant tPTEN−/− mice compared with an age-matched WT control (C57BL/6 background). (C) Quantitative RT-PCR analysis of Traf6 expression in 146a1 and 146b transgenic mice compared with littermate controls on WT and tPTEN−/− backgrounds. (D) Western blot analysis of Stat1 protein expression in total thymocytes and mature T cells from 12-week-old littermate 146a8 transgenic and WT mice. (E) Western blot analysis of Traf6 expression, IκBα degradation, and c-myc induction in TCR-stimulated total thymocytes from 10-week-old 146a1/tPTEN−/− and 146b/tPTEN−/− mice and littermate or age-matched tPTEN−/− controls. Thymocytes were stimulated with PMA and ionomycin for the indicated times. Data in (A-C) are presented as mean ± SD and are representative of 3 independent experiments. Statistics were calculated by Student t test (***P < .001, **P < .01, *P < .05, n.s. not significant).
In retrovirally transduced mature T cells, miR-146a has been shown to attenuate TCR signaling by targeting Traf6, a ubiquitin ligase that plays a key role in activating NF-κB.20,22 We hypothesized that miR-146 might act similarly in thymocytes to dampen the TCR signals that drive TCRα/c-myc translocations. Indeed, we found that Traf6 expression is reduced in tPTEN−/− thymocytes from premalignant and tumor-bearing mice (Figure 5B). In addition, in both mir-146a and mir-146b transgenic mice, we observed a significant reduction in Traf6 expression at both the transcript and protein levels on WT and tPTEN−/− backgrounds (Figure 5C and supplemental Figure 4). The reduction in Traf6 expression was not caused by global downregulation of miR-146 target genes because the expression of Stat1, a reported target of miR-146a in T cells whose 3′ UTR has only partial homology to the miR-146a seed sequence,23 was not downregulated in either thymocytes or mature T cells of the mir-146a transgenic mice (Figure 5D). We next tested whether Traf6 downregulation by miR-146 results in decreased NF-κB activation downstream of TCR signals by measuring degradation of the NF-κB inhibitor IκBα. Upon phorbol 12-myristate 13-acetate (PMA) and ionomycin stimulation, we observed that IκBα degradation was significantly attenuated in mir-146 transgenic thymocytes (Figure 5E and supplemental Figure 4). At later time points, we also observed attenuated induction of the NF-κB target gene c-myc (Figure 5E and supplemental Figure 4). Because TCRα/c-myc translocations in tPTEN−/− premalignant thymocytes are dependent on intact TCR signaling, miR-146 attenuation of TCR signaling might reduce the incidence of these translocations, explaining the delay in c-myc upregulation we observe. In addition, diminished endogenous c-myc levels caused by NF-κB inhibition might increase the threshold for survival and proliferation of transforming thymocytes.
Reduced proliferative capacity of mir-146b transgenic CD4 T cells
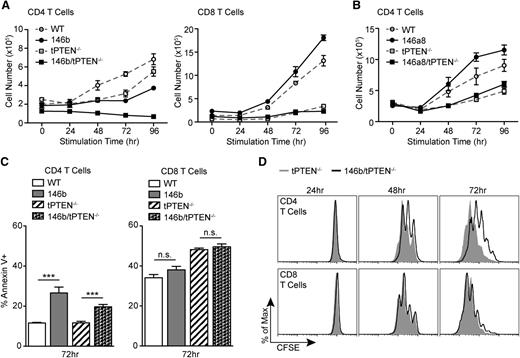
As we have shown before, T-cell development in mir-146a and mir-146b transgenic mice is largely normal, with the exception of reduced CD4 T cells in the mir-146b transgenic mice. Because TCR signals are important for survival of mature T cells,24 we wondered whether miR-146b attenuation of TCR signaling might contribute to the lower T-cell numbers. Strikingly, we found that mir-146b transgenic CD4 T cells were significantly impaired in their ability to expand in response to TCR stimulation (Figure 6A). This effect was amplified on the tPTEN−/− background. Co-cultured CD8 T-cell expansion was normal, likely because of the absence of transgenic mir-146b expression (Figure 6A). Surprisingly, mir-146a transgenic CD4 T-cell expansion was completely normal (Figure 6B). This observation is in agreement with our finding that mir-146a transgenic mice have normal T-cell proportions.
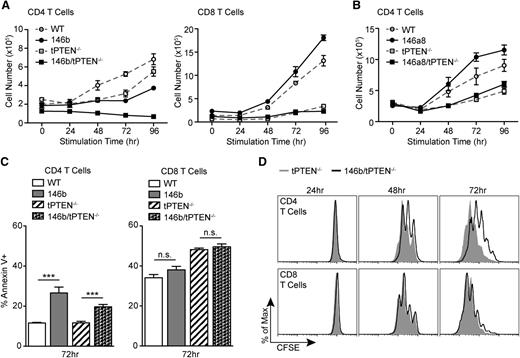
miR-146b negatively regulates CD4 T-cell proliferation. (A) TCR-mediated expansion of CD4 and CD8 T cells from 146b transgenic mice and littermate controls on WT and tPTEN−/− backgrounds. Total T cells were stimulated with anti-CD3/CD28. Triplicate T-cell samples were collected at each time point and total live T-cell numbers were determined by Trypan blue exclusion. CD4 and CD8 numbers were calculated based on flow cytometric analysis at each time point. Data are representative of 4 independent experiments. (B) TCR-mediated expansion of CD4 T cells from 146a8 transgenic mice and littermate controls on WT and tPTEN−/− backgrounds. The assay was performed as described in (A). Data are representative of 2 independent experiments, one for each founder line. (C) Quantification of flow cytometric analysis of Annexin V+ T cells stimulated as in (A) for 72 hours. Data are representative of 2 independent experiments. (D) Flow cytometric analysis of CFSE dilution over time by T cells labeled with 1 μM CFSE before stimulation as described in (A). Histograms are representative of duplicate samples and 2 independent experiments. All mice used were littermate pairs between 6 and 10 weeks old (C57BL/6). Data in (A-C) are presented as mean ± SD and statistics were calculated by Student t test (***P < .001, n.s. not significant).
miR-146b negatively regulates CD4 T-cell proliferation. (A) TCR-mediated expansion of CD4 and CD8 T cells from 146b transgenic mice and littermate controls on WT and tPTEN−/− backgrounds. Total T cells were stimulated with anti-CD3/CD28. Triplicate T-cell samples were collected at each time point and total live T-cell numbers were determined by Trypan blue exclusion. CD4 and CD8 numbers were calculated based on flow cytometric analysis at each time point. Data are representative of 4 independent experiments. (B) TCR-mediated expansion of CD4 T cells from 146a8 transgenic mice and littermate controls on WT and tPTEN−/− backgrounds. The assay was performed as described in (A). Data are representative of 2 independent experiments, one for each founder line. (C) Quantification of flow cytometric analysis of Annexin V+ T cells stimulated as in (A) for 72 hours. Data are representative of 2 independent experiments. (D) Flow cytometric analysis of CFSE dilution over time by T cells labeled with 1 μM CFSE before stimulation as described in (A). Histograms are representative of duplicate samples and 2 independent experiments. All mice used were littermate pairs between 6 and 10 weeks old (C57BL/6). Data in (A-C) are presented as mean ± SD and statistics were calculated by Student t test (***P < .001, n.s. not significant).
A defect in TCR-mediated T-cell expansion could be caused by insufficient activation through the TCR. However, mir-146b transgenic CD4 T cells upregulated activation markers CD69 and CD25 normally (supplemental Figure 5). Alternatively, this defect could be caused by increased apoptosis. In response to TCR stimulation, we observed a modest increase in apoptosis of 146b CD4 T cells (Figure 6C). However, we were unable to rescue the expansion defect with the pan-caspase inhibitor zVAD-FMK (supplemental Figure 6), suggesting that increased cell death is not the critical defect in 146b CD4 T cells. Interestingly, we found that 146b CD4 T cells have a pronounced defect in proliferation, with a reduced ability to dilute carboxyfluorescein succinimidyl ester (CFSE) over time (Figure 6D). Cell-cycle analysis of BrdU-pulsed T cells revealed that this defect was caused by an inhibition of the G0/G1 to S-phase cell-cycle transition (24 hours and 48 hours, Figure 7A). In mature T cells, NF-κB activation is important for inducing expression of genes that drive the G0/G1 to S-phase transition, most importantly c-myc.25 Interestingly, Traf6 expression was significantly decreased in 146b/tPTEN−/− and to some extent 146a/tPTEN−/− CD4 T cells (Figure 7B). Consistent with a corresponding reduction in NF-κB activity, we found that c-myc levels were significantly reduced in 146b CD4 T cells and to a lesser extent in 146a CD4 T cells (Figure 7C and supplemental Figure 7). Furthermore, increased expression of the cell-cycle inhibitor p27 in naïve 146b CD4 T cells and somewhat in 146a CD4 T cells was observed. Thus, decreased c-myc induction and high p27 levels likely cooperate to inhibit 146b T-cell proliferation.
miR-146b negatively regulates the G0/G1 to S-phase cell-cycle transition by targeting Traf6. (A) BrdU vs 7AAD flow cytometric analysis of T cells stimulated with anti-CD3/CD28 antibodies and pulsed with 10 μM BrdU for 1 hour before harvest at the indicated time points. Plots are representative of duplicate samples. (B) Western blot analysis of Traf6 protein expression in purified CD4 T cells stimulated with anti-CD3/CD28 antibodies for the indicated times. (C) Western blot analysis of c-myc induction and p27 levels in purified CD4 T cells stimulated with anti-CD3/CD28 antibodies for the indicated times. Mice in (A-C) were littermate pairs between 6 and 8 weeks old on the C57BL/6 background (146a8/tPTEN−/− in [B] was age matched). Data in (A-C) are representative of 2 independent experiments.
miR-146b negatively regulates the G0/G1 to S-phase cell-cycle transition by targeting Traf6. (A) BrdU vs 7AAD flow cytometric analysis of T cells stimulated with anti-CD3/CD28 antibodies and pulsed with 10 μM BrdU for 1 hour before harvest at the indicated time points. Plots are representative of duplicate samples. (B) Western blot analysis of Traf6 protein expression in purified CD4 T cells stimulated with anti-CD3/CD28 antibodies for the indicated times. (C) Western blot analysis of c-myc induction and p27 levels in purified CD4 T cells stimulated with anti-CD3/CD28 antibodies for the indicated times. Mice in (A-C) were littermate pairs between 6 and 8 weeks old on the C57BL/6 background (146a8/tPTEN−/− in [B] was age matched). Data in (A-C) are representative of 2 independent experiments.
Discussion
Inactivation of the tumor suppressor PTEN renders cells vulnerable to malignant transformation; however, additional oncogene activation is frequently necessary to drive forward the transformation process.4 In human T-ALL, PTEN loss is frequently coupled with increased expression of the c-myc oncogene.26 Activating mutations of NOTCH1, which are found in 50% of T-ALL patients, also drive concurrent c-myc amplification and PTEN suppression.8,27 In mice, c-myc upregulation is observed in both T cell–specific and hematopoietic-specific PTEN-deficient mice,11,28,29 largely driven by the presence of TCRα/c-myc chromosomal translocations.11,30 In one model, concurrent deletion of c-myc completely abrogated leukemia/lymphoma development.28 Thus, in both humans and mice, activation of c-myc plays a key role in promoting development of PTEN-deficient T-cell malignancies. Here, through analysis of premalignant lesions in tPTEN−/− mice, we have identified 2 miRNAs that act to delay this crucial c-myc induction and inhibit development of lymphomas.
Through analysis of mir-146a and mir-146b T cell–specific transgenic mice, we found that both miRNAs act to suppress TCR signals in thymocytes. Intact TCR signaling is a prerequisite for TCRα/c-myc translocations in PTEN-deficient premalignant thymocytes.11 By dampening TCR signals, these miRNAs might act to decrease the incidence of translocations, thus delaying c-myc upregulation and thymocyte transformation. Although much of what drives chromosomal translocations between particular genetic loci remains unclear, recent data suggest that translocations occur more frequently in genic regions that are expressed vs silent.31 By targeting Traf6, miR-146 represses NF-κB–dependent transcription of c-myc downstream of the TCR and might thereby decrease the participation of the c-myc locus in translocations. Alternatively, reduced endogenous c-myc expression might increase the threshold for survival and proliferation of cells that already contain genomic aberrations.
The perfect seed sequence homology between miR-146a and miR-146b predicts that these 2 miRNAs should have the same mRNA targets and function redundantly.18 Consistent with this, our studies indicate that both miRNAs act similarly to repress Traf6, thereby negatively regulating NF-κB signaling and c-myc induction. However, the magnitude of this phenotype was somewhat attenuated in mir-146a vs mir-146b transgenic mice. The impact of this was most apparent in mature T cells, where transgenic expression of mir-146b, but not mir-146a, resulted in decreased CD4 T-cell numbers and impaired TCR-mediated proliferation. However, although this T-cell phenotype has not been previously reported for miR-146b, retroviral overexpression of miR-146a in T cells was shown to impair TCR-mediated proliferation. Conversely, mir-146a deficiency promotes hyperproliferation of T cells in response to antigen.20 Hence there is likely a threshold of miR-146 activity necessary to disrupt normal T-cell function. Nevertheless, miR-146a and miR-146b extend tPTEN−/− survival to a similar extent, which implies that in the context of tumor formation and growth, the more modest degree of miR-146a activity is sufficient to mediate its antitumor effects. Thus, increased miR-146 expression might slow expansion of malignant CD4 T cells or premalignant thymocytes in tPTEN−/− mice. In further support of redundant function of these miRNAs, we also found that mir-146a deficiency alone is not sufficient to accelerate lymphomagenesis in tPTEN−/− mice (supplemental Figure 8).
Traf6 is a key activator of NF-κB downstream of TCR signaling.22 Consistent with Traf6 being a primary target of miR-146 in thymocytes and T cells, miR-146 overexpression phenocopies aspects of NF-κB deficiency in T cells, which results in decreased T-cell numbers and/or impaired TCR-mediated proliferation and survival.25,32-36 This defect was attributed to inhibition of NF-κB–dependent induction of c-myc, resulting in defective entry into S-phase.25 Similarly, in mir-146b transgenic CD4 T cells, we observed reduced c-myc induction and delayed S-phase entry in response to TCR stimulation. Thus, attenuation of NF-κB signaling through targeting of Traf6 likely accounts for much of the mir-146b mature T-cell phenotype. More modest targeting of Traf6 by miR-146a and differential regulation of p27 levels likely account for the normal phenotype of mir-146a transgenic T cells.
Our data support a tumor-suppressive role for miR-146a and miR-146b in murine T-cell malignancies mediated by repression of TCR signaling and NF-κB activation. These observations are supported by recent analysis of mir-146a–deficient mice, which develop a multiorgan inflammatory disease starting at 6 to 8 months of age that culminates in the development of myeloid sarcomas and some B- and T-cell lymphomas.37,38 Consistent with miR-146 playing a tumor-suppressive role in human myeloid malignancies, miR-146a was significantly downregulated in several studies profiling miRNA expression in acute and chronic myeloid leukemia (AML/CML) samples and myelodysplastic 5q– syndrome.39-43 Also, enforced expression of miR-146a in AML cells lines inhibited cell proliferation and increased sensitivity to antileukemic drugs.41 In support of a tumor-suppressive role for miR-146 in human T-cell malignancies, a comprehensive screen of miRNA expression in 50 primary T-ALL samples showed considerable downregulation of miR-146a expression in T-ALL samples compared with normal T-cell controls.44 Interestingly, studies have shown that NF-κB is often activated downstream of NOTCH1 in human T-ALL cell lines, and treatment with the NF-κB inhibitor bortezomib significantly inhibits their growth.45,46 Hence, miR-146–based therapeutics that act to downmodulate NF-κB activity might be effective in treating T-ALL. In support of this notion and its potentially broader applicability, reexpression of miR-146a in cancer cell lines of nonhematopoietic tumor types in which NF-κB is hyperactivated, such as pancreatic and breast cancer, has been shown to repress NF-κB activity and reduce proliferative and invasive capacity.47,48
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Lin He for insightful discussions, David Baltimore for miR-146a−/− mice, and Heather Melichar and Margaux Bennett for critical review of the manuscript.
This work was supported by National Institutes of Health, National Cancer Institute grant CA168007 and a CRCC fellowship (M.L.B.), and was supported in part by National Institutes of Health, National Cancer Institute grant CA66236 (A.W.).
Authorship
Contribution: M.L.B., L.X., and A.W. conceived and designed the experiments; M.L.B., L.X., Y.S., and C.K. performed the experiments; and M.L.B. and A.W. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Astar Winoto, University of California, Berkeley, Department of Molecular & Cell Biology,142 Life Sciences Addition #3200, Berkeley, CA 94720; e-mail: winoto@berkeley.edu.
References
Author notes
M.L.B. and L.X. contributed equally to this study.


![Figure 7. miR-146b negatively regulates the G0/G1 to S-phase cell-cycle transition by targeting Traf6. (A) BrdU vs 7AAD flow cytometric analysis of T cells stimulated with anti-CD3/CD28 antibodies and pulsed with 10 μM BrdU for 1 hour before harvest at the indicated time points. Plots are representative of duplicate samples. (B) Western blot analysis of Traf6 protein expression in purified CD4 T cells stimulated with anti-CD3/CD28 antibodies for the indicated times. (C) Western blot analysis of c-myc induction and p27 levels in purified CD4 T cells stimulated with anti-CD3/CD28 antibodies for the indicated times. Mice in (A-C) were littermate pairs between 6 and 8 weeks old on the C57BL/6 background (146a8/tPTEN−/− in [B] was age matched). Data in (A-C) are representative of 2 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/26/10.1182_blood-2013-11-539411/4/m_4089f7.jpeg?Expires=1769189538&Signature=1aV3ZCXTJEoNgqj5mmY-cXZTata8ErBNE5rrYtYfrGeRfiLtjGCBZU-uhHu6Jm2iLLT-vsV0STI3y7CU1cKkZuEPgy8Snia2yJel7JN0r-~ifnDBh6wUYAT3sBIRRsYQbUMu1ndoB2xCEGkCkS-ktEw~hn81Lkf0Hvf3qNrnOEZ7lqjgVHcFWeXkAG0lZ8-6Kbp2pJPB0kO45-THWjQW-lcRJNRRMwM-0e3~1utSG7Hv662VSAOpyNnthHwMmiU36KuojafjIG-~gpLu8nNisIV4yewqnTgBbLQwh~MvkjO4FEVdoZi68oHds7E8T1kbso7mlQEAXYrCX~IQGGzGDw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 7. miR-146b negatively regulates the G0/G1 to S-phase cell-cycle transition by targeting Traf6. (A) BrdU vs 7AAD flow cytometric analysis of T cells stimulated with anti-CD3/CD28 antibodies and pulsed with 10 μM BrdU for 1 hour before harvest at the indicated time points. Plots are representative of duplicate samples. (B) Western blot analysis of Traf6 protein expression in purified CD4 T cells stimulated with anti-CD3/CD28 antibodies for the indicated times. (C) Western blot analysis of c-myc induction and p27 levels in purified CD4 T cells stimulated with anti-CD3/CD28 antibodies for the indicated times. Mice in (A-C) were littermate pairs between 6 and 8 weeks old on the C57BL/6 background (146a8/tPTEN−/− in [B] was age matched). Data in (A-C) are representative of 2 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/26/10.1182_blood-2013-11-539411/4/m_4089f7.jpeg?Expires=1769518630&Signature=OiYVzLSAiUdUqD6pniIdpXcv2SAMwZnLE2u0QxSPOl--kpZDiXvmZ0ERsIcp6W278M1cbyojKcbl7Sn9r8Kqur4-e8N9RBxMRKo5IpGX8GMDAxCiIMWjPvd8442LKMl9VmmZePUpXBqk6yY99a6xTYc3-nvGIUw5Ls5j0m0qnYMrq-~ASGV-ku9lb1U29H4AAtuo36GGs0zbLeGDDs-ZjEmrHVvtLAuA9zcxxoO~6JCyhyk5ugN5BO4YWnyV9Z0H-~fKS5vewBzh60reBa~muW-qw9W9CFHoqXGZXX988r0Vl3kbhXmkuhht7Kgg6X2JJwqnS0HrUuPuGje8vHmosw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)