In this issue of Blood, Mraz et al show that microRNA-150 (miR-150) is the most abundantly expressed miR in chronic lymphocytic leukemia (CLL) and affects the threshold for B-cell receptor (BCR) signaling by repressing expression levels of GAB1 and FOXP1. This functional link might explain the described association between expression levels of miR-150 and prognosis.1
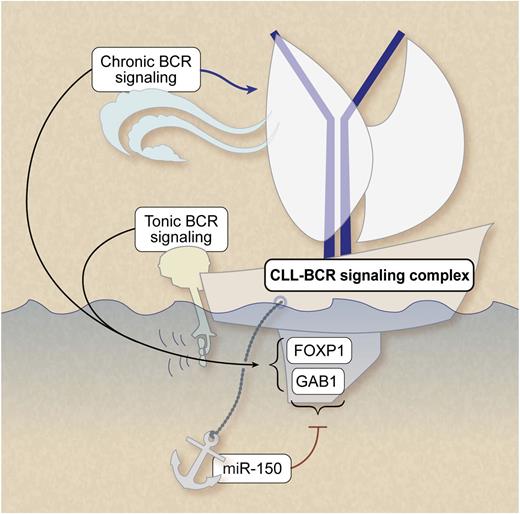
Ligand-independent (“tonic”) and ligand-dependent (“chronic”) BCR signaling play a pivotal role in CLL survival and growth. MiRNA-150 dampens the threshold for BCR signaling by repressing expression levels of GAB1 and FOXP1. Professional illustration by Debra T. Dartez.
Ligand-independent (“tonic”) and ligand-dependent (“chronic”) BCR signaling play a pivotal role in CLL survival and growth. MiRNA-150 dampens the threshold for BCR signaling by repressing expression levels of GAB1 and FOXP1. Professional illustration by Debra T. Dartez.
In 2002, the first link between small noncoding RNAs, known as miRs, and cancer was made by the observation that in CLL, the most common genetic aberration 13q14 deletion, was associated with downregulation of miR-15a and miR-16-1, which reside in the minimally deleted region within 13q14.2 This seminal observation initiated many studies into the role of miRs in the pathogenesis of cancer in general and especially in CLL. The pioneering work on miR-15a and 16-1, however, also exemplified the complexity of aberrant miR expression and its possible relation with alterations in cancer-specific biological pathways. Although early studies suggested that in CLL miR-15a/16-1 mediated control of BCL2 expression and survival, it took until 2010 to learn that in fact the function of these miRs in B-cell malignancies is exerted mainly by downregulation of genes controlling cell-cycle entry.3
Because each miR can affect the expression of hundreds of different genes, which not only differs per cell type but also depends on their developmental stage, and because currently available bio-informatic tools are imperfect in predicting targets via sequence similarities, it has been highly challenging to interpret the pathophysiological relevance of aberrations in miR levels measured ex vivo, or after in vitro manipulation.
Despite these challenges, over the years, several miRs could convincingly be mapped to disease-specific relevant pathways, such as the identification of miR-34a as a component of the chemotherapy resistance network in CLL.4
Major advances have been made in understanding the molecular pathogenesis underlying CLL progression and treatment resistance, demonstrating a pivotal role for ligand-independent (“tonic”) and ligand-dependent (“chronic”) BCR signaling in CLL trafficking and activation-induced interaction with the tumor microenvironment. Recent studies in model systems also demonstrated that miRs are involved in BCR signaling.5 In CLL, a strong hint for a role of specific miRs in the susceptibility to BCR signaling came from observations on differential expression levels of approximately 20 miRs in immunoglobulin heavy-chain variable subgenes (IGHV) unmutated and/or ζ-associated protein of 70-kDa (ZAP-70)–positive CLL cases vs mutated IGHV and/or ZAP-70 negative cases (reviewed in Mraz and Kipps 6 ). However, for most of these miRS, their exactly relevant target genes in CLL have not yet been identified and therefore their exact role in the BCR signaling pathway is still largely unclear.
The detailed work of Mraz et al in this issue of Blood takes this next step. They functionally map the most abundantly expressed miR in CLL, miR-150, to the BCR signaling pathway via 2 specific target genes.1 First, they show that miR-150 levels were significantly lower in cases that used unmutated IGHV or that expressed ZAP-70. To identify miR-150 target genes in CLL, transcriptome analysis was performed and data were verified using 6 database tools. Two genes with evolutionary conserved binding sides for miR-150 were identified, namely GAB1 and FOXP1. The link was functionally validated in cell lines and primary CLL samples by transfection studies. Moreover, roles for both miR-150 and the 2 target genes were shown by measuring calcium fluxes after BCR ligation and RNA interference in cell lines. The BCR-induced calcium fluxes correlated with miR-150 levels in primary CLL samples. Importantly, they also showed that miR-150 levels are significantly lower in cases that use unmutated IGHV or that express ZAP-70. Moreover, patient survival studies indicate an association between expression levels of miR-150, GAB1, FOXP1, and clinical outcome. Although expression levels of miR-150 correlated with ZAP-70 or IGHV status, low-level expression of miR-150 had an independent prognostic value for both overall survival and treatment-free survival.
GAB1 and FOXP1 are involved in essential signaling cascades in both normal and malignant B cells. GAB1 is an adaptor molecule of phosphoinositide 3-kinase (PI3K) (reviewed in Zhang et al7 ). Because the PI3K pathway is activated not only by the BCR, but also by a variety of receptor tyrosine kinases and cytokine receptors, this signaling route is pivotal in microenvironment-induced CLL activation. FOXP1 is a transcription factor associated with the activated B-cell phenotype of diffuse large B-cell lymphoma, and it has been suggested that truncated forms of FOXP1 are functionally associated with subtypes of diffuse large B-cell lymphoma characterized by constitutive nuclear factor-κB activity.8 It would be of interest to learn the functional consequences of miR-mediated regulation of these genes in the context of microenvironmental-induced features of activation such as survival and chemosensitivity, proliferation, and adhesion/migration. In this respect, the observations in model systems that stimulation with anti-CD40 antibodies largely prevented BCR-mediated changes in miR expression levels5 indeed indicate an intricate interplay with miRs, BCR signaling, and microenvironmental stimuli.
The importance of BCR signaling in the pathobiology of CLL is underscored by the significant clinical activity of inhibitors blocking BCR-associated kinases, specifically Bruton tyrosine kinase and PI3K. Because these need to be administered for prolonged periods, balance of costs vs effectiveness of these novel drugs is highly important. Predictive markers of effectiveness are therefore highly needed. Data provided by Mraz et al suggest that miR-150 expression and its target genes might influence the sensitivity of malignant B cells to these inhibitors.
As always, after a step forward has been taken, we quickly wonder about the implications. One question to consider is: Why and how is miR-150 regulated? If low miR-150 levels facilitate BCR signaling, which is advantageous to the cells, we might expect selection for this state. Indeed, Mraz et al find that miR-150 levels negatively correlate with disease progression (Rai stage). As for regulation, the authors provide a hint that miR-150 might be controlled epigenetically. Finally, an exciting new field is the development of miR-based therapeutics that either downregulate the function of oncogenic miRs or upregulate the expression of tumor-suppressive miRs, such as the liposome-based miR-34 mimic currently in phase 1 trials (reviewed in Ling et al9 ). Because Mraz et al identified miR-150 as a potential key regulator of PI3K and because it has been showed in follicular B cells that miR-185 regulates the expression of Bruton tyrosine kinase,10 it does not take a lot of imagination to speculate on the development of miR mimetics to target key signaling cascades in CLL.
Conflict-of-interest disclosure: The authors declare no competing financial interests.