Key Points
B cells but not red cells are GPI deficient in PIGM-associated IGD, caused by a core promoter mutation that abrogates Sp1 binding.
In red but not B cells, PIGM transcription is independent of Sp1 binding to the core promoter, hence GPI expression in red cells is near normal.
Abstract
A rare point mutation in the core promoter −270GC-rich box of PIGM, a housekeeping gene, disrupts binding of the generic transcription factor (TF) Sp1 and causes inherited glycosylphosphatidylinositol (GPI) deficiency (IGD). We show that whereas PIGM messenger RNA levels and surface GPI expression in IGD B cells are low, GPI expression is near normal in IGD erythroid cells. This divergent phenotype results from differential promoter chromatin accessibility and binding of Sp1. Specifically, whereas PIGM transcription in B cells is dependent on Sp1 binding to the −270GC-rich box and is associated with lower promoter accessibility, in erythroid cells, Sp1 activates PIGM transcription by binding upstream of (but not to) the −270GC-rich box. These findings explain intact PIGM transcription in IGD erythroid cells and the lack of clinically significant intravascular hemolysis in patients with IGD. Furthermore, they provide novel insights into tissue-specific transcriptional control of a housekeeping gene by a generic TF.
Introduction
Hypomorphic coding mutations in genes of the glycosylphosphatidylinositol (GPI) biosynthetic pathway cause inherited GPI deficiency (IGD).1-6 Unique to PIGM-associated IGD is a homozygous hypomorphic mutation (−270C>G) in the core promoter of this mannosyltransferase-encoding gene.7,8 PIGM transcription in B cells is dependent on the generic transcription factor (TF) Sp1, and in IGD B cells, the pathogenic mutation abrogates binding of Sp1 to its cognate −270GC-rich motif.8,9 This leads to histone hypoacetylation and polycomb-dependent PIGM transcriptional repression and thus to IGD8,9 Patients with PIGM-associated IGD suffer from life-threatening splanchnic vein thrombosis and intractable epilepsy but not clinically significant intravascular hemolytic anemia.7 The latter, caused by lack of the GPI-linked complement regulators CD55 and CD59 from the surface of erythrocytes, is a defining feature of paroxysmal nocturnal hemoglobinuria (PNH),10,11 a disorder caused by somatic mutations in PIGA in hematopoietic stem cells and their progeny.12,13 Although the −270C>G mutation in PIGM-associated IGD is constitutional, the degree of IGD is variable and differs between different cell types, a feature shared with the other forms of IGD.1-5,7 In particular, despite its marked deficiency in IGD B cells, expression of GPI in erythroid cells in PIGM-associated IGD and presumably in other forms of IGD is largely preserved.
Here, we investigate the transcriptional mechanisms that underpin the blood cell type-specific differences in GPI expression in PIGM-associated IGD.
Patients and methods
The patient with IGD and the heterozygous parents were previously described.7 Blood samples were obtained and stored under approval by the local research ethics committees; verbal informed consent was provided in accordance with the Declaration of Helsinki.
Detailed methods are described in the supplemental Methods available on the Blood Web site.
Results and discussion
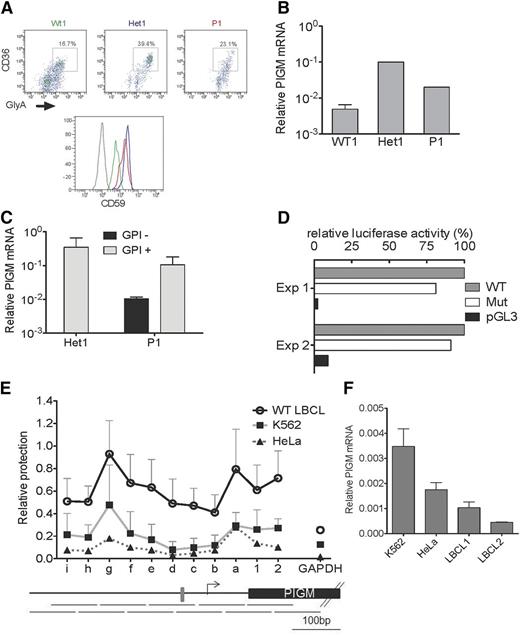
Although surface expression of GPI in primary PIGM IGD red blood cells is near normal (>90% GPI positive), >70% of primary B cells (supplemental Figure 1) and >90% of patient-derived lymphoblastoid B cell lines (LBCLs) are GPI negative.9 To address whether differential GPI expression reflects differences in transcriptional regulation of PIGM, we first assessed GPI and PIGM messenger RNA (mRNA) levels in primary B and erythroid cells from a patient with IGD. In line with the near-normal GPI expression in mature erythrocytes, we found that cell-surface GPI levels (as assessed by anti-CD59 staining) and PIGM mRNA in erythroid precursors from a patient with IGD (P1) and his heterozygous parent (Het1) were actually higher than in normal control cells (Figure 1A-B). In contrast, PIGM mRNA levels in sorted GPI-negative primary B cells from P1 were lower than in their GPI-positive counterparts and Het1 B cells (Figure 1C). In line with intact PIGM transcription in erythroid precursors, in the erythroid K562 cell line, a 2-kb PIGM promoter fragment with the C>G mutation was nearly as active as the corresponding WT fragment (Figure 1D).
PIGM transcription and chromatin accessibility in erythroid and B cells. (A) Fluorescence-activated cell sorter analysis of cell-surface expression of CD59 in primary erythroid precursor cells from a normal wild-type (WT) donor (green), heterozygous (Het1; blue) parent, and patient with IGD (P1; red). Erythroid precursor cells were generated from peripheral blood mononuclear cells, and flow cytometric analysis was performed on day 7 of the culture. Histograms show CD59 expression on erythroid precursors identified as CD36+GlyA+ cells (upper panel). Gray line: isotypic control. (B) PIGM mRNA expression in normal, Het1, and P1 primary erythroid precursor cells. PIGM mRNA levels were assessed by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) in flow-sorted CD36+GlyA+ erythroid precursor cells as shown in (A). Data were normalized to GAPDH. Het1 and P1, n = 1; normal, n = 4. (C) PIGM mRNA expression in Het1 and P1 B cells. PIGM mRNA levels were assessed by qRT-PCR in sorted primary CD19+GPI+ and CD19+GPI− B cells (see supplemental Figure 1A). GPI expression was assessed after staining with flourescent aerolysin. Note that heterozygous B cells are all GPI+ . Data were normalized to GAPDH. Het1, n = 2; P1, n = 2. (D) WT and mutated (Mut) PIGM promoter transcriptional activity in vitro. Luciferase reporter assays were performed in K562 cells. Cell lysates were assayed for luminescence 48 hours after transfection. Luminescence values were normalized to renilla, and results are presented relative to the activity of the WT promoter construct. pGL3 represents a promoter-less control construct. Mean values of 2 independent experiments, each performed in triplicate assays, are shown. (E) Nucleosome mapping by MNase protection assay at the length of the PIGM promoter in WT LBCL, HeLa, and K562 cells. The abundance of eleven 100-bp amplicons that overlap by 50 bp (shown below the graph) was quantified by quantitative polymerase chain reaction using equal amounts of MNase-digested or undigested DNA, as described the supplemental Methods. The gray box indicates the −270GC-rich box. Letters represent amplicons upstream of the transcription start site (TSS; shown as an arrow), whereas numbers denote regions downstream of the TSS. (F) PIGM mRNA expression in 2 WT LBCL, HeLa, and K562 cells. PIGM mRNA levels were assessed by qRT-PCR. Data were normalized to GAPDH and are shown as mean ± standard error of the mean (n = 3).
PIGM transcription and chromatin accessibility in erythroid and B cells. (A) Fluorescence-activated cell sorter analysis of cell-surface expression of CD59 in primary erythroid precursor cells from a normal wild-type (WT) donor (green), heterozygous (Het1; blue) parent, and patient with IGD (P1; red). Erythroid precursor cells were generated from peripheral blood mononuclear cells, and flow cytometric analysis was performed on day 7 of the culture. Histograms show CD59 expression on erythroid precursors identified as CD36+GlyA+ cells (upper panel). Gray line: isotypic control. (B) PIGM mRNA expression in normal, Het1, and P1 primary erythroid precursor cells. PIGM mRNA levels were assessed by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) in flow-sorted CD36+GlyA+ erythroid precursor cells as shown in (A). Data were normalized to GAPDH. Het1 and P1, n = 1; normal, n = 4. (C) PIGM mRNA expression in Het1 and P1 B cells. PIGM mRNA levels were assessed by qRT-PCR in sorted primary CD19+GPI+ and CD19+GPI− B cells (see supplemental Figure 1A). GPI expression was assessed after staining with flourescent aerolysin. Note that heterozygous B cells are all GPI+ . Data were normalized to GAPDH. Het1, n = 2; P1, n = 2. (D) WT and mutated (Mut) PIGM promoter transcriptional activity in vitro. Luciferase reporter assays were performed in K562 cells. Cell lysates were assayed for luminescence 48 hours after transfection. Luminescence values were normalized to renilla, and results are presented relative to the activity of the WT promoter construct. pGL3 represents a promoter-less control construct. Mean values of 2 independent experiments, each performed in triplicate assays, are shown. (E) Nucleosome mapping by MNase protection assay at the length of the PIGM promoter in WT LBCL, HeLa, and K562 cells. The abundance of eleven 100-bp amplicons that overlap by 50 bp (shown below the graph) was quantified by quantitative polymerase chain reaction using equal amounts of MNase-digested or undigested DNA, as described the supplemental Methods. The gray box indicates the −270GC-rich box. Letters represent amplicons upstream of the transcription start site (TSS; shown as an arrow), whereas numbers denote regions downstream of the TSS. (F) PIGM mRNA expression in 2 WT LBCL, HeLa, and K562 cells. PIGM mRNA levels were assessed by qRT-PCR. Data were normalized to GAPDH and are shown as mean ± standard error of the mean (n = 3).
These findings suggest that the −270GC-rich motif is required for PIGM transcriptional activity in B cells but not in erythroid cells.
To investigate whether the differential role of the −270GC-rich motif in PIGM transcription in erythroid vs B cells entails differences in promoter accessibility, we mapped nucleosome occupancy by nucleosome protection assay in the core and proximal promoter of PIGM in K562 cells and LBCLs (Figure 1E). We found that in B cells, nucleosome occupancy at the promoter region spanning the −270 motif is higher compared with K652 and epithelial HeLa cells in which the core promoter is nearly nucleosome-free. These findings are specific to PIGM, because nucleosome occupancy at the core promoter of the housekeeping gene GAPDH was uniformly low in all 3 cell types. Next, we addressed whether these differences impact on transcriptional activity. We found that PIGM mRNA levels in LBCLs were lower than in K562 and HeLa cells (Figure 1F), in keeping with higher nucleosome occupancy at the PIGM promoter in B cells and suggesting that chromatin configuration at the promoter of PIGM is less permissive for transcription in B cells than other cell types. Given these differences in transcriptional potential, we propose that the pathogenic C>G mutation is sufficient to tip the promoter into a repressive state in B but not erythroid cells, thereby in part explaining the differential cellular phenotype in PIGM-associated IGD.
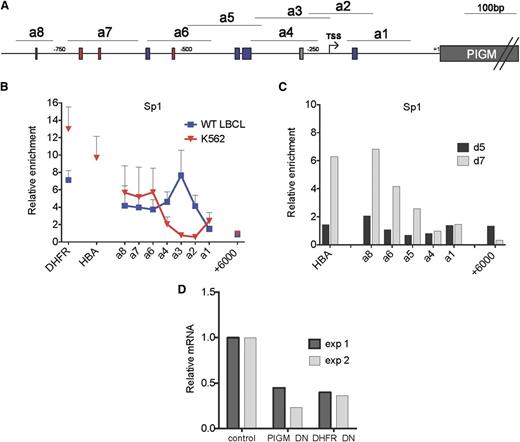
Next, we considered whether lineage-specific differences in the binding of the generic TF Sp1 at the promoter of PIGM are responsible for, or contribute to, active transcription of PIGM and intact GPI biosynthesis in IGD erythroid cells. As before, in B cells,9 we found high levels of Sp1 binding not only at the −270GC-rich box but also in upstream promoter regions where 3 GC-rich boxes are predicted to bind Sp1 (Figure 2A-B). By contrast, in K562 cells, we observed high levels of Sp1 binding corresponding to the upstream GC-rich boxes but not at the −270GC-rich box (Figure 2B). We further confirmed lack of binding of Sp1 to the −270GC-rich box in primary erythroid cells (Figure 2C). To further establish the functional role of Sp1 in PIGM transcription, we introduced a dominant-negative form of Sp114 into K562 cells. This led to a >50% reduction in PIGM mRNA levels (Figure 2D), suggesting that Sp1 is required for active PIGM transcription in erythroid cells. Therefore, Sp1 regulates PIGM transcription in erythroid cells by binding to upstream, but not to the core promoter GC-rich boxes, providing a mechanism for the intact PIGM transcription and near-normal GPI expression in IGD erythroid cells.
Differential binding of Sp1 to the −270GC-rich box in erythroid and B cells. (A) Schematic of PIGM promoter. a1 to a8 are amplicons of the PIGM promoter used in chromatin immunoprecipitation analysis. The gray box denotes the position of the experimentally validated Sp1 binding site mutated in IGD at position −270 from ATG. Red and blue boxes correspond to predicted GATA1 and Sp1 binding sites, respectively. (B) Sp1 binding at the length of the PIGM promoter in WT LBCL and K562 cells. DHFR and α-globin represent amplicons in the promoter region of the respective genes and are shown as positive controls. +6000bp represents an amplicon 6 kbp downstream of the ATG start codon of PIGM and is shown as a negative control. n = 5 to 6 independent experiments. (C) Sp1 binding at the PIGM promoter in primary erythroid precursor cells. Sp1 binding was assessed in d5 and d7 cord blood–derived erythroid precursors by chromatin immunoprecipitation quantitative polymerase chain reaction. Results are presented as fold enrichment over immunoglobulin G control (n = 1). (D) The impact of a dominant-negative (DN) form of Sp1 on PIGM transcription in K562 cells. Cells were transduced either with a green fluorescent protein (GFP)-expressing retrovirus containing a complementary DNA encoding a DN form of Sp1 or with a no-insert GFP control. mRNA levels were assessed by qRT-PCR in flow-sorted purified cells 48 hours later and normalized against GAPDH. The graph shows PIGM and DHFR mRNA levels in cells transduced with the GFP-DN Sp1 construct relative to GFP-only control. Two independent experiments are shown.
Differential binding of Sp1 to the −270GC-rich box in erythroid and B cells. (A) Schematic of PIGM promoter. a1 to a8 are amplicons of the PIGM promoter used in chromatin immunoprecipitation analysis. The gray box denotes the position of the experimentally validated Sp1 binding site mutated in IGD at position −270 from ATG. Red and blue boxes correspond to predicted GATA1 and Sp1 binding sites, respectively. (B) Sp1 binding at the length of the PIGM promoter in WT LBCL and K562 cells. DHFR and α-globin represent amplicons in the promoter region of the respective genes and are shown as positive controls. +6000bp represents an amplicon 6 kbp downstream of the ATG start codon of PIGM and is shown as a negative control. n = 5 to 6 independent experiments. (C) Sp1 binding at the PIGM promoter in primary erythroid precursor cells. Sp1 binding was assessed in d5 and d7 cord blood–derived erythroid precursors by chromatin immunoprecipitation quantitative polymerase chain reaction. Results are presented as fold enrichment over immunoglobulin G control (n = 1). (D) The impact of a dominant-negative (DN) form of Sp1 on PIGM transcription in K562 cells. Cells were transduced either with a green fluorescent protein (GFP)-expressing retrovirus containing a complementary DNA encoding a DN form of Sp1 or with a no-insert GFP control. mRNA levels were assessed by qRT-PCR in flow-sorted purified cells 48 hours later and normalized against GAPDH. The graph shows PIGM and DHFR mRNA levels in cells transduced with the GFP-DN Sp1 construct relative to GFP-only control. Two independent experiments are shown.
We also tested the role of GATA1, a megakaryocyte-erythroid TF,15 and of KLF1,16,17 an erythroid-specific TF that shares the same GC-rich binding motifs with the Sp TF family.18,19 We found high-level binding of GATA-1 at the PIGM promoter in K562 cells (supplemental Figure 2A) but not in primary erythroid cells (supplemental Figure 2B). KLF-1, in primary erythroid precursors (KLF1 is not expressed at protein level in K562 cells20 ), binds highly to the upstream PIGM promoter regions but not to the −270GC-rich box (supplemental Figure 2C). These data show that whereas GATA1 and KLF1 are likely to be important for PIGM transcription, their function is independent of the −270GC motif and thus the C>G mutation would not be expected to impact on their ability to regulate PIGM transcription in IGD erythroid cells.
Taken together, our data provide insights in 3 areas. First, they provide a molecular basis for the genotype-phenotype divergence between erythroid and B cells in patients with PIGM-associated IGD. However, the presence of a small fraction of GPI-negative red blood cells and GPI-positive B cells (supplemental Figure 1) in PIGM-associated IGD suggests that the differential binding of Sp1 to the −270GC-rich box in these cells might not be absolute. Perhaps in a stochastic manner, Sp1 occasionally fails to sufficiently activate PIGM transcription in IGD erythroid cells, whereas in a minority of primary B cells, either effective binding of Sp1 to the mutated motif or a yet to be defined compensatory transcriptional process activates PIGM transcription. Our findings also explain how defective GPI biosynthesis, a defining feature for both PNH and PIGM-associated IGD, leads to clinically significant intravascular hemolysis in the former but not the latter. Finally, they show that lineage-specific transcriptional regulation of an active gene might be determined by differential chromatin accessibility and binding of a generic TF to different areas of the promoter, thus highlighting a novel mechanism of gene transcription.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by Fundação para a Ciência e Tecnologia, Portugal (J.R.C.), the Medical Research Council (V.S.C.), the Sir Halley Stewart Trust (K.M.), and the National Institute for Health Research Biomedical Research Centre at Imperial College Healthcare NHS Trust.
Authorship
Contribution: J.R.C., K.M., and V.S.C. performed experiments and interpreted data, and A.K., I.A.G.R., A.M.A., and D.M.L. conceived and supervised the work and wrote the paper with input from all authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Anastasios Karadimitris, Centre for Haematology, Department of Medicine, Imperial College London, Hammersmith Hospital, Du Cane Rd, London W12 0NN, UK; e-mail: a.karadimitris@imperial.ac.uk.