Abstract
Genome-wide association studies (GWAS) have identified common susceptibility loci in developing chronic lymphocytic leukemia (CLL). As the price on whole exome and genome sequencing continues to drop, large sequencing studies become more feasible and will change our understanding of the biological fundament of malignancy - and of the premalignant state of monoclonal lymphocytosis toward leukemic status. However, CLL is heterogeneous and, while co-inheritance of multiple low-risk variants govern predisposition, GWAS cannot yet be expected to contribute sufficiently in delineating individual molecular drivers. There is a need to be able to determine likely drivers in single patients, while GWAS continue to add information to draw from. We hypothesized that with the right tool much information can be extracted from the patient specific analysis - with direct implications in the clinic. The production cost is already within range of other lab analyses, but the interpretative work is still in its infancy in the clinical settings.
AIM: We set out to engineer a method to automatically rank and annotate whole exome data efficiently, pointing to most likely contributing variants in the development of MBL and CLL - with direct practical implications for the clinic and research.
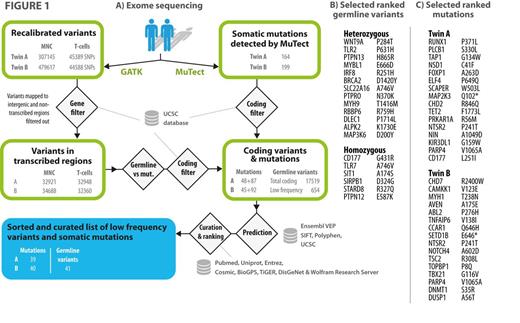
METHODS: As testing ground and model for the tool we analysed a pair of identical twins in whom one had developed B-CLL and the other monoclonal lymphocytosis (notably with different clonal usage of light chains). In this setting extensive variant and mutation annotation, filtering, analysis and ranking were carried out through variant calling and MuTect output post-processing tool written in the versatile Wolfram programming language (Mathematica 9, Wolfram Research, Oxfordshire, UK) tied to external data sources (Fig. 1). Variants are automatically evaluated and ranked on the basis of 1) allele frequency, predicted damage, non-synonymous change (e.g. charge or polarity change), affected region or predicted size of frameshift, 2) functional annotation of gene and 3) automated literature search and gene-disease association. 4) Querying COSMIC database determines calculated mutational frequency (normalized to size) of the affected gene in human cancers and whether the gene is known to be involved in oncogenesis. 5) Finally, graphical representation of expected expression is provided to assist interpretation.
RESULTS: Rare inherited variants in the B-Cell Receptor and Wnt/beta-catenin Signaling Pathway and tumor suppressors were found in the germline set (Fig. 1). These results also showed IRF8 mutation, recently linked to CLL susceptibility loci, and a reported susceptibility loci (rs17246404). Twin A could be distinguished from B by a series of high-ranked mutations with myeloid association (RUNX1, TET2, PLCB1 and ELF4 etc, fig. 1C), a nonsense mutation affecting MAP2K3 and a frameshifting indel in CHST15 - a gene suggested to act as a B-cell receptor important for B-cell development. Also, a missense mutation with altered charge in the helicase domain of CLL-associated CHD2 was found. Twin B was found to have a different set of relevant mutations that may also be drivers in inducing a proliferative disorder (e.g. ABL2 SH2 domain, MYH1 motor domain, TNFAIP6 CUB domain, NOTCH4 EGF-like calcium-binding domain). Twin B, although following a benign course, had an overall higher number of mutations was backed by affected genes involved in DNA repair response (TOPBP1 and DNMT1). Also a key feature was chromatin, histone and DNA modifiers (CHD7, SETD1B and likewise DNMT1).
DISCUSSION: The use of a new set of scripts allowed for the easy interpretation of variants in these twins, in whom it can be surmised that master genes underlying the B-cell proliferation in B-CLL can be found. We suggest that this analysis method can be used for individual assessment in close collaboration or directly by the clinician. As example, primary refractoriness to chemotherapy is an evident problem, but much data is already at hand to evaluate potential problematic variants. In our case doxorubicin and imatinib influx transporters were reported as possibly affected. There is a need for constructing analysis methods that assist the clinician in evaluating the disease on a genomic level. Hopefully, this kind of approach will make similar ventures in the single patient a clinical reality within a short span of time - benefitting patient and clinics.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.