Abstract
Background:
Ring sideroblasts (RS) are erythroblasts with iron-loaded mitochondria, appears as blue granules when stained with Prussian blue, and are characteristic of myelodysplastic syndromes (MDS) subgroups refractory anemia with ring sideroblasts (RARS), refractory cytopenia with multilineage dysplasia and ring sideroblasts (RCMD-RS), and RARS with thrombocytosis (RARS-T) but are also infrequently found in other MDS groups. The etiology of RS remains unclear. Molecular genetic analysis of MDS patients with RS (MDS-RS) using exome and targeted sequencing identified mutations in several genes including SF3B1. While the RS phenotype was shown to be strongly associated with SF3B1 mutations, the association of SF3B1 mutations with better prognosis remained controversial. Also, there remained several patients who do not show any mutations in SF3B1 suggesting there are additional genes associated with RS phenotype. In addition to SF3B1 mutations, RARS-T patients show mutations in JAK2 (V617F and Exon 12), MPL and CALR genes, frequently mutated in Essential Thrombocytopenia (ET). But there remained a high number of patients who do not show any mutation in JAK2, MPL and CALR genes suggesting additional genes are associated with thrombocytosis. In order to gain further insight into the molecular genetics of MDS-RS patients and to elucidate their clinical significance we screened a large cohort of patients with RS (RARS/RCMD-RS) and a subgroup with thrombocytosis (RARS-T).
Methods:
This study is approved by the Institutional Review Board of Columbia University and informed consent was obtained from all the individuals participated in the study. Bone marrow mono nuclear cells were isolated from bone marrow aspirate and DNA was extracted using DNeasy Blood and Tissue kit. To screen for most frequent mutations in SF3B1 (exon 14 and 15), MPL (exon 10), JAK2 (exon 12), and CALR (exon 9) genes, primers were designed to amplify the exons and the exon-intron junctions using PCR and the amplified and purified PCR products were sequenced using Sanger sequencing on both strands. To screen for mutations V617F in JAK2, primers were designed to carry out allele specific PCR. Demographic and clinical data is collected from patient’s charts/reports. Statistical analysis including survival curve (Kaplan–Meier method) was performed using GraphPad Prism Software.
Results:
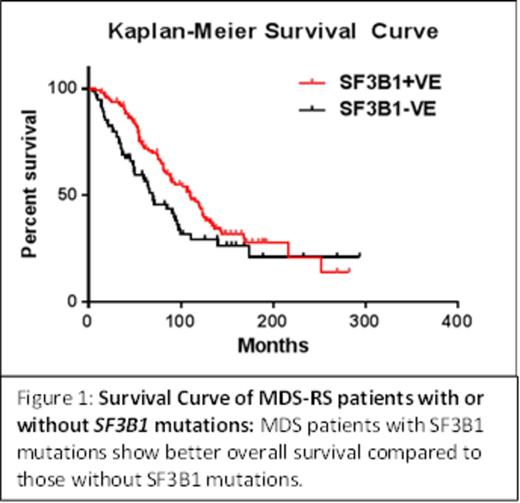
A total of 209 patients with RS phenotype were screened for SF3B1 mutations. Mutations in SF3B1 gene were detected in 62% (130/209) of RS patients, including 85% (23/27) of RARS-T and 59% (107/182) of RARS/RCMD-RS patients. Among all the SF3B1 mutations, K700E was the most frequent mutation (60%) followed by mutations at H662. Clinical significance of SF3B1 mutations on overall survival, using Kaplan-Meier method, showed SF3B1 mutations were associated with better prognosis (Fig 1). The median survival of patients with SF3B1 mutation is 110 months compared to those without mutations, 70 months (p value < 0.034). Studies to screen SF3B1 negative patients for additional mutations are being carried out using exome sequencing to identify genes associated with RS phenotype.
In addition to SF3B1, RARS-T patients were also screened for JAK2 V617F and exon 12 mutations. JAK2 V617F mutations were detected in 11% of RARS-T patients and no mutations were found in JAK2 exon 12. Patients negative for JAK2 were screened for mutations in MPL and CALR genes. No mutations were found in MPL but 8% (2/23) of those negative for JAK2 were found to have CALR mutations. Both CALR mutations were frameshift mutations that alter the C-terminus thus abolishing the KDEL sequence required for CALR function. There are still 80% RARS-T patients in our cohort who do not show any mutations in JAK2, MPL, CALR genes.
Conclusions:
The association of SF3B1 mutation with prognosis is controversial with some studies suggesting a better prognosis while other reports no effect. Our study, using a large cohort of well-characterized RS patients provides an Independent verification of the observation that SF3B1 mutations are associated with better overall survival. There remains a large group of patients (40% in RARS/RCMD-RS groups) without SF3B1 mutation but with a RS phenotype suggesting yet other gene(s) is associated with RS phenotype. Similarly, there remain patients who do not show any mutations in JAK2, MPL and CALR suggesting other genes are involved in the etiology of thrombocytopenia.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.