Abstract
Introduction: Canonical notion demonstrates that Cell fate is determined by transcriptional factor. Accordingly, the lineage specific transcriptional factors have been investigated for various kinds of cells. Especially, in the hematopoietic system, the extensive research for lineage specific transcriptional factors had elucidated the transcriptional factors which regulated lineage commitment. B cell commitment and development requires the activities of multiple transcription factors, including the early B cell factor (EBF), PAX5, and E2A. These transcription factors regulate B cell development in a stage specific manner. The hematopoietic progenitor cells which are deficient for any of them, cannot commit B cells. Among them EBF1 is presumed to be more potent. Rescue early pro B cell to induce the expression of several key proteins including RAG that enable gene rearrangement to occur by opening of IgH locus.
We found that the B cell developmental arrest caused by EBF1 deficiency can be rescued by a single non coding RNA. These B cells which are deficient EBF1 but showed the expression of CD19, B cell lineage specific surface marker and VDJ recombination, molecular markers of B cell commitment in vitro B cell differentiation system cocultured with Tst4 cells, stromal cell lines. We further investigated the quality and differentiation potential of these B lineage commitment cells in the in vivo mouse model and elucidated the mechanism of this phenomenon.
Material and methods: We collected EBF1−/− fetal liver hematopoietic progenitor (Lin−) cells and cultured them on TSt-4 stromal cells after infecting with non coding RNA and control vector in IMDM medium containing cytokines and then injected it into NOG mice. Collect bone marrow (BM), thymus and spleen from those mice. Then comprehensive Gene-Expression analysis, real time PCR for VDJ recombination analysis was performed and checked surface marker by flowcytometry.
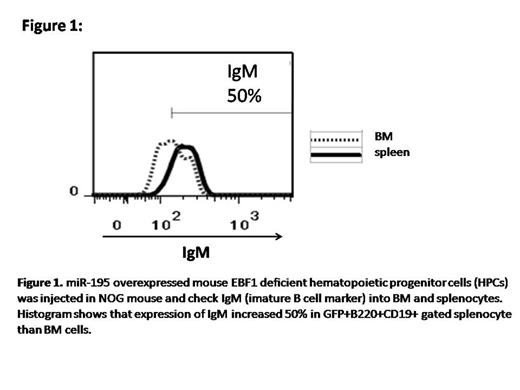
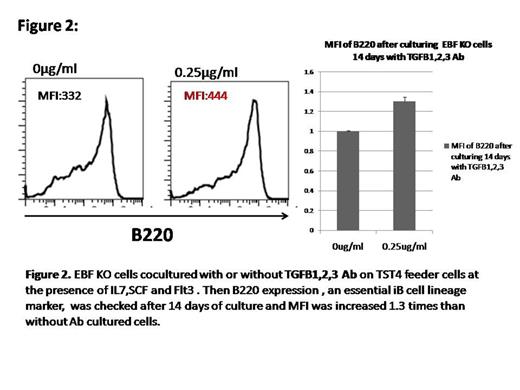
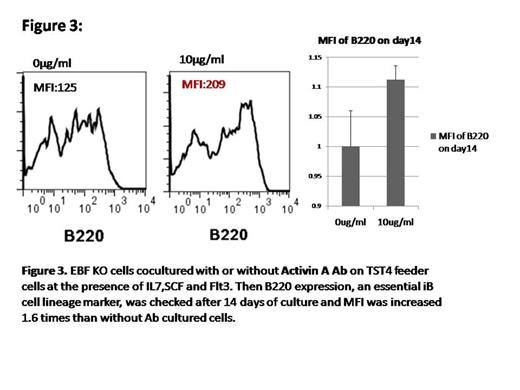
Result: We analyzed BM and spleen of non coding RNA infected EBF1 KO cells injected mice and found the expression of CD19 in BM as well as in spleen and upregulated of B220 also, comparing with control vector expressed cells. Furthermore, surface IgM expression of CD19 positive cells in the spleen is upregulated compared with the cells in the BM (Figure 1). Several target genes of the non-coding RNAs were identified by use of cDNA array analysis and luciferase reportor assay. Among them, several genes were involved in TGF beta pathway. As TGF beta family and the pathway, has been reported a critical factor which is negatively regulating B lymphopoiesis (Figure 2). We hypothesized that TGF beta family genes such as Tgfβr3, Acvr2a, are responsible for B cell differentiation for which EBF1 is dispensable .We cultured EBF KO cells for 14 days with and without TGF beta 1,2,3 antibody and Activin A antibody on TST4 cells. We found that increase mean intensity (MFI) of B220 into antibodies positive cocultured cells (Figure 3) to suggest, the suppression of TGF beta pathway is partially responsible for B cell differentiation under EBF1 deficiency.
Conclusion: Canonical notion of cell fate determination of B cells defines that EBF1 is an indispensible factor for B lymphopoiesis. However, from our previous and present study it is proved that without EBF1 B cell development can progress to pro B to pre B cell and Immature B cell stage and “VDJ recombination” occur in the absence of EBF. Furthermore, in vivo mouse model, EBF1 deficient hematopoietic progenitor cells differentiated into IgM positive cells. Therefore we can conclude that EBF is dispensable of VDJ recombination, opening of IgH locus, binding of RAG protein and B cell differentiation to the mature stage. One of the mechanisms is possibly due to the stimuli from microenvironment, such as TGF beta family and pathway. Furthermore, the detail mechanism of IgH locus opening, epigenetic changes and chromatin remodeling around the IgH locus in the absence of EBF is under investigation.
Chanda:Japan Society for the Promotion of Science(JSPS): Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.