Key Points
hSWI/SNF requires WASp to remodel IFNG and TBX21 loci in T-helper (TH)1 cells.
WAS-causing but not XLT-causing hot-spot mutations impair SWI/SNF-activity at TH1 gene promoters.
Abstract
Wiskott-Aldrich syndrome (WAS), an immunodeficiency disorder, and X-linked thrombocytopenia (XLT), a bleeding disorder, both arise from nonsynonymous mutations in WAS, which encodes a hematopoietic-specific WASp. Intriguingly, XLT evolves into WAS in some patients but not in others; yet the biological basis for this cross-phenotype (CP) effect remains unclear. Using human T-helper (TH) cells expressing different disease-causing WAS mutations, we demonstrated that hSWI/SNF-like complexes require nuclear-WASp to execute their chromatin-remodeling activity at promoters of WASp-target, immune function genes during TH1 differentiation. Hot-spot WAS mutations Thr45Met and Arg86Cys, which result in XLT-to-WAS disease progression, impair recruitment of hBRM- but not BRG1-enriched BAF complexes to IFNG and TBX21 promoters. Moreover, promoter enrichment of histone H2A.Z and its catalyzing enzyme EP400 are both impaired. Consequently, activation of Notch signaling, a hBRM-regulated event, and its downstream effector NF-κB are both compromised, along with decreased accessibility of nucleosomal DNA and inefficient transcription-elongation of WASp-target TH1 genes. In contrast, patient mutations Ala236Gly and Arg477Lys that manifest in XLT without progressing to WAS do not disrupt chromatin remodeling or transcriptional reprogramming of TH1 genes. Our study defines an indispensable relationship between nuclear-WASp– and hSWI/SNF-complexes in gene activation and reveals molecular distinctions in TH cells that might contribute to disease severity in the XLT/WAS clinical spectrum.
Introduction
WASp, the protein mutated in Wiskott-Aldrich syndrome (WAS) supports actin polymerization in the cytoplasm by verprolin-homology/cofilin-homology/acidic(VCA)-domain using an actin-related-protein (ARP)2/3-dependent mechanism,1 and transcription in the nucleus through a mechanism that is undefined, but appears to be ARP2/3-independent.2,3 WAS mutations are causative of the human disease,4 and are linked to at least 3 clinical phenotypes (ie, gene pleiotropy).5 Loss-of-function mutations associate with X-linked thrombocytopenia (XLT), a platelet defect presenting with bleeding symptoms, or WAS, where, in addition to thrombocytopenia, immunodeficiency, autoimmunity, and cancer predominate.6-9 In WAS, multiple cell-lineages of the immune system are affected.10 Gain-of-function mutations associate with X-linked neutropenia (XLN), a white-cell defect presenting with infections.11 Loss-of-function missense mutations that still allow expression of mutant WASp are enigmatic in that the same mutation is associated with either XLT or XLT that progresses to WAS. Yet predicting which XLT patient will or will not develop WAS is impossible. Notably, WAS genotype-phenotype correlations show a predilection for missense mutations occurring in WASp-homology1(WH1)-domain but not VCA-domain to manifest XLT-to-WAS disease progression.6-9 Such observations suggest novel, VCA:ARP2/3-independent function(s) of WASp in immunobiology, whose impairment we postulate contributes to the development of XLT-to-WAS cross-phenotype (CP) progression.5
Elucidating the underlying mechanism for the CP effect in WAS is a major challenge in the field. Currently, neither XLT nor XLT-to-WAS progression can be modeled in mice. Therefore, one approach to elucidate the pathophysiologic mechanism underlying CP effects is to perform functional studies of gene mutations directly in human cells that are relevant to WAS symptomatology.5 Because immune dysregulation is a primary manifestation of human WAS,6,8 we investigated the molecular effects of WAS mutations on human CD4+T-helper(TH) lymphocytes crucial for adaptive immunity.
We previously identified a nuclear location of WASp in TH1 cells, where it is required for the “activating” histone H3K4me3 modification at the TBX21 promoter, a TH1-network gene that is also a WASp-target gene.2 Furthermore, nuclear-WASp but not cytoplasmic-WASp associates with RBBP5 (a subunit of mixed lineage leukemia [MLL]-complex) and catalyzes H3K4me3 modification independently of its VCA-domain, and this domain, we show, is nonessential for TH1 gene activation.3 In Xenopus oocytes, the gene activation function of Wave1, a WASp-family protein, is also independent of its VCA-like (verprolin-homology) domain.12 Although these findings align well with the observation that VCA-domain missense mutations generally do not result in XLT-to-WAS progression in humans,6-9 the question remains: why are some WAS mutations more pathogenic to the immune system whereas others spare it completely? Answering this will require clarifying the mechanism(s) by which WASp functions in the nucleus of human TH cells and other immune cells, and how individual WAS mutations differ in their proclivity to perturb this process.
Here, we addressed both issues, focusing mainly on the chromatin events of RNA polymerase II–dependent transcription, because the newfound nuclear roles of WASp, Neural-WASp, and Wave1 (all WASp family proteins) are linked to transcription.2,12,13 Our study uncovers an unanticipated relationship between WASp and hSWI/SNF-complex in the TH1 cell nucleus. It identifies 2 mechanistic, disease-related signaling cascades, WASp→BRM→Notch→NF-κB and WASp→EP400→H2A.Z, whose differential impairment by WH1-mutations, but not non–WH1-mutations, correlates with clinical severity grades in human XLT/WAS disease spectrum.
Methods
Cells
Peripheral blood mononuclear cells from a normal donor and a WAS patient were used to establish CD4+ TH-cell lines by transformation with human T-lymphotropic virus type-1 (HTLV-1) as described.14 Immortalized WASnull TH cells were transfected to express different disease-causing mutations.3 Briefly, full-length (FL)-WASp cDNA was subcloned into mammalian expression vector pCMV6 containing Flag/Myc dual-tags (Origene). All disease-associated, amino acid substitutions in WASp cDNA were performed using QuickChangeIISite-Directed Mutagenesis (Stratagene) (supplemental Table 1, available on the Blood Web site), and mutant sequences were confirmed by DNA sequencing. FL-WASp and its mutants were transfected into Jurkat or WASnull TH cells by AmaxaNucleofector (Lonza). Stable expression was verified by immunoblotting and flow cytometry. Cells were cultured under TH1- or TH2-skewing or nonskewing TH0 conditions as described.3
Mass spectrometry
Multiple MS assays on immunoprecipitated WASp-enriched protein complexes were performed as described.3 Briefly, lysates from micrococcal nuclease (MNase)-treated nuclei of human primary or Jurkat TH1-skewed cells expressing WASp-Flag/Myc dual-tagged protein were incubated with anti-WASp, or anti-Flag and anti-Myc, or their isotype-Ig antibodies. The recovered polypeptides were analyzed by nano-LC-mass spectrometry (MS)/MS on an LTQ-Orbitrap Velos mass spectrometer using the SEQUEST database.
Coimmunoprecipitation and immunoblotting
Flow cytometry
Intracellular protein staining was performed as described.3 Briefly, cells were treated with protein transport inhibitors (GolgiPlug and GolgiStop), T-cell receptor (TCR)-activated, fixed, permeabilized, and labeled with fluorochrome-conjugated antibodies against cytokines and/or transcription factors. Cells were analyzed with a Becton-Dickinson LSRII using FACSDiva. Geometric mean fluorescence intensity was calculated using quadrant/histogram statistics.
Enzyme-linked immunosorbent assay
The supernatant was harvested from untransfected and WASp mutant transfected TH0 (nonskewed) or TH1- or TH2-skewed cells, and normal TH cells in culture. Expression of granulocyte macrophage–colony-stimulating factor, interferon gamma (IFNG), and interleukin 4 (IL4) cytokines was quantitated by enzyme-linked immunosorbent assay (ELISA) (R&D) in 3 replicates.
Chromatin IP (ChIP)-quantitative polymerase chain reaction and reverse-transcriptase polymerase chain reaction
All single- and sequential-ChIP assays were performed with MNase-digested chromatin isolated from ∼5000 cells after fixing protein-DNA interactions with 1% formaldehyde as described.2,3 ChIP-grade antibodies and their isotype-Ig antibodies were used to pull-down DNA:Protein complexes (supplemental Table 1). ChIPed samples were used for quantitative polymerase chain reaction (qPCR) analysis and the derived Ct values converted to absolute copy numbers using cloned-DNA plasmid standard dilution curve. Nonspecific signals obtained with IgG-ChIP were subtracted from test samples. RNA prepared from ∼5000 nonskewed TH0, TH1-, or TH2-skewed cells using Quick-RNA MiniPrep (Zymo Research) was used to synthesize cDNA, and the reverse transcriptase PCR (RT-PCR) analysis performed on a 7500HT RT-PCR System (Applied Biosystems) using primers/probes (detailed in supplemental Table 1). RT-PCR and ChIP-qPCR were performed from the same biological sample. The derived Ct-values were converted to absolute copy numbers with a cloned-DNA plasmid standard dilution curve2,3
Quantitative DNase I hypersensitivity (DHS) assay
The PCR-based DHS assay was performed as described.2 Briefly, T-cell nuclei were isolated with the Nuclei Isolation Kit (Sigma-Aldrich) and incubated with 6 U of DNase I enzyme and 1 μg of nuclei at 37°C for 10 minutes. This dose was selected as optimum from a serial dilution curve performed previously.2 Samples were analyzed by qRT-PCR to determine the number of amplicons remaining after nuclease treatment.
Electrophoretic mobility shift assay
Nuclear and cytosolic fractions were isolated from TH cells. Electrophoretic mobility shift assay (EMSA) was carried out in 5 g nuclear extract and oligonucleotide-containing consensus for NF-κB binding site. Oligonucleotide was end-labeled to a specific activity of 105 CPM using γ-[32P]-ATP and T4-polynucleotide kinase and purified on a Nick column. Reaction mixtures with radiolabeled oligonucleotides were incubated for 20 minutes and then resolved on 6% nondenaturing polyacrylamide gels after adding 0.1% bromophenol blue, and then subjected to autoradiography. For competition assays, 30-fold excess unlabeled oligonucleotide (competitor) or unlabeled mutated oligonucleotide (Mutator) was added for 20 minutes after incubation with oligonucleotides. For super-shift assays, specific antibody was added for 20 minutes after the reaction with oligonucleotide was completed.
Results
Nuclear-WASp associates with hSWI/SNF-components
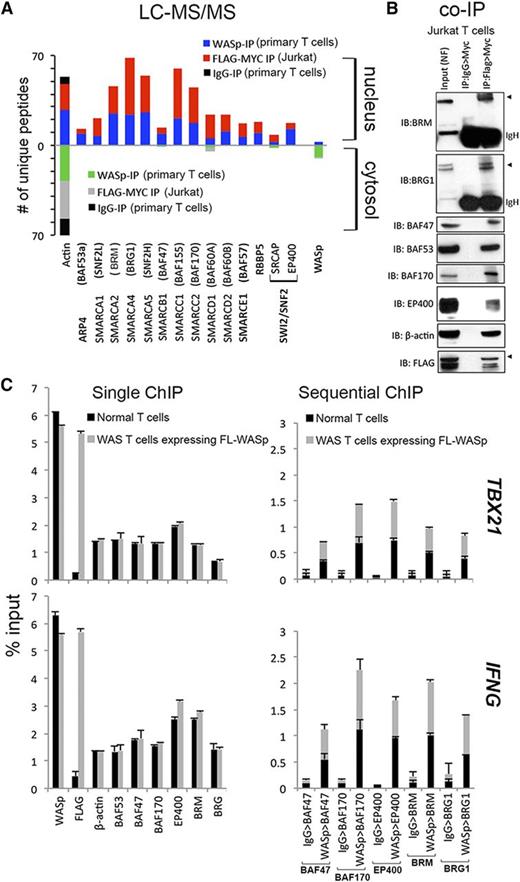
Gene promoters that lack WASp or Wave1 show impaired H3K4me3 enrichment,2,12 which in the case of WASp occurs consequent to MLL (RBBP5) perturbation.2 Because SWI/SNF-like chromatin-remodeling complexes (CRCs) favor binding H3K4me3-marked promoters,15 we hypothesized that WASp effect on transcription is linked to MLL-SWI/SNF chromatin-signaling module. To this end, we queried our mass-spectrometry (MS) proteomic dataset of nuclear and cytoplasmic WASp complexes previously generated from human TH1-skewed cells, both primary and Jurkat T cells, the latter transfected with Flag/Myc-tagged WASp.3 Using a candidate, hypothesis-driven approach, we captured polypeptides of hSWI/SNF-complexes (hBRM, BRG1, BAF47, BAF53a, BAF57, BAF60, BAF155, BAF170, SMARCA1, and SMARCA5), hSWR1 (EP400, SRCAP), and β-actin from both endogenous WASp and Flag/Myc-tagged transfected WASp (Figure 1A and supplemental Figure 1). The presence of hBRM and the absence of BAF180 from the WASp-proteome reported by MS (Figure 1A) suggest that nuclear-WASp associates predominantly with BAF but not PBAF complexes in human TH1-skewed cells.16-18 Notably, only nuclear-WASp but not cytoplasmic-WASp associates with CRCs. Although the binding of some of the CRCs to nuclear-WASp could be nonspecific, the persistence of these associations after MNase digestion favors the capture of specific chromatin-located, WASp:SWI/SNF-complexes, as previously demonstrated for other chromatin-bound proteins.19,20 Nonetheless, the WASp:SWI/SNF association yielded by MS was validated by sequential coIP (1stIP:anti-Flag; 2ndIP:anti-Myc) in MNase-treated Jurkat TH1-skewed cells transfected with Flag/Myc doubly-tagged WASp (Figure 1B). The collective presence of these SWI/SNF subunits suggests a role for nuclear-WASp in remodeling the gene locus with which it interacts.
Characterizing physiologic WASp:SWI/SNF associations in vivo. (A) Mass spectrometry. Actual number of polypeptides of WASp-associated, chromatin-remodeling complexes (CRC) captured from cytosolic and nuclear fractions of TH1-skewed, primary TH (endogenous WASp) or Jurkat TH (Flag/Myc-tagged transfected WASp) analyzed by LC-MS/MS after immunoaffinity purification with anti-WASp, anti-FLAG/MYC (sequential 2-step purification), or -IgG antibodies, as described.3 Searches from 3 to 4 biological replicates were combined to generate a MultiConsensus report of peptides and proteins identified from the WASp proteome after applying the filtering criteria previously described3 (supplemental Figure 1). (B) Selective validation of MS-generated WASp-associated CRC proteome by co-IP. Protein complexes isolated by 2-step IP (1st:Flag, 2nd:Myc) from the nuclear fraction of TCR-activated, TH1-skewed, Jurkat TH stably expressing Flag/Myc-tagged WASp were resolved by sequential western blotting with the same gel with indicated antibodies. This image is part of the full gel image shown in Figure 2D. (C) MNase ChIP-qPCR. Chromatin enrichment profiles of the indicated proteins, at 5′UTR (promoter region) of the indicated genes in TH1-skewed, normal, or WASnull TH cells stably transfected with Flag/Myc-tagged, full-length (FL) WASp. For sequential ChIP, 2 rounds of conventional ChIPs were performed in the indicated sequence (eg, WASp>BAF47 denotes 1stChIP:WASp, 2ndChIP:BAF47; IgG>BAF47 denotes1stChIP:IgG, 2ndChIP:BAF47). The displayed ChIP values (mean ± SEM) are percentages of total nuclear input chromatin and were derived after subtracting the background values obtained with isotype IgG antibody, the latter not shown for single ChIPs. Data were generated from 3 biological replicates. The genomic location of PCR primer/probes is shown in Figure 2C.
Characterizing physiologic WASp:SWI/SNF associations in vivo. (A) Mass spectrometry. Actual number of polypeptides of WASp-associated, chromatin-remodeling complexes (CRC) captured from cytosolic and nuclear fractions of TH1-skewed, primary TH (endogenous WASp) or Jurkat TH (Flag/Myc-tagged transfected WASp) analyzed by LC-MS/MS after immunoaffinity purification with anti-WASp, anti-FLAG/MYC (sequential 2-step purification), or -IgG antibodies, as described.3 Searches from 3 to 4 biological replicates were combined to generate a MultiConsensus report of peptides and proteins identified from the WASp proteome after applying the filtering criteria previously described3 (supplemental Figure 1). (B) Selective validation of MS-generated WASp-associated CRC proteome by co-IP. Protein complexes isolated by 2-step IP (1st:Flag, 2nd:Myc) from the nuclear fraction of TCR-activated, TH1-skewed, Jurkat TH stably expressing Flag/Myc-tagged WASp were resolved by sequential western blotting with the same gel with indicated antibodies. This image is part of the full gel image shown in Figure 2D. (C) MNase ChIP-qPCR. Chromatin enrichment profiles of the indicated proteins, at 5′UTR (promoter region) of the indicated genes in TH1-skewed, normal, or WASnull TH cells stably transfected with Flag/Myc-tagged, full-length (FL) WASp. For sequential ChIP, 2 rounds of conventional ChIPs were performed in the indicated sequence (eg, WASp>BAF47 denotes 1stChIP:WASp, 2ndChIP:BAF47; IgG>BAF47 denotes1stChIP:IgG, 2ndChIP:BAF47). The displayed ChIP values (mean ± SEM) are percentages of total nuclear input chromatin and were derived after subtracting the background values obtained with isotype IgG antibody, the latter not shown for single ChIPs. Data were generated from 3 biological replicates. The genomic location of PCR primer/probes is shown in Figure 2C.
WASp and SWI/SNF complexes coenrich at TH1-gene promoters in vivo
To test the aforementioned hypothesis, we focused mainly on “core” TH1-genes (IFNG, TBX21), because nuclear localization of WASp and its targeting to “core” TH2 genes (IL4, GATA3) are negligible in TH2-skewed cells (supplemental Figure 2A).3 Full-length (FL), Flag/Myc-tagged WASp was stably expressed in a patient-derived TH-line lacking endogenous WASp (previously described).3 Upon TH1-skewing, ChIP demonstrates that endogenous CRCs (BAF47, BAF53, BAF170, hBRM, BRG1, EP400) and transfected Flag-WASp (or endogenous WASp) are both enriched at the promoters of IFNG and TBX21 (Figure 1C). WASp (or Flag) is not enriched at the promoter of CSF2, a WASp-nontarget gene in TH1-skewed cells, or at COL8A2-TRAPPC3 intergenic locus that is devoid of protein-coding genes (supplemental Figure 2A), which together denote targeting specificity of WASp in TH1-cell genome. Significantly, sequential-ChIP showed that WASp and CRCs co-occupy the same promoter regions of IFNG and TBX21 genes (Figure 1C).
Deficient TH1-gene expression is linked to WAS-causing, WH1-domain mutations but not with XLT-causing, non-WH1 mutations
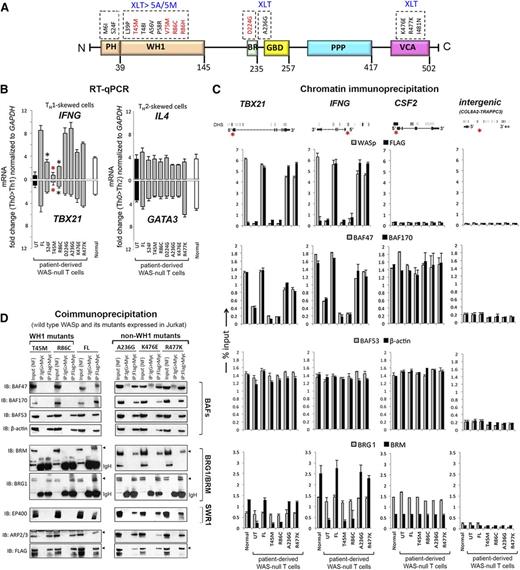
Because CRC activity is essential for remodeling TH1-gene loci,16,21-23 we asked whether pathogenic WAS missense mutations impair TH1 gene induction and, if so, whether they do so by disrupting CRC function at TH1 gene promoters. We introduced disease-causing, point mutations in full-length WASp and stably expressed each mutant in patient-derived TH cells lacking endogenous WASp. Mutations in PH-domain (S24F), WH1-domain (T45M, R86C), basic-domain (A236G, D224G), and VCA-domain (K476E, R477K) all reported to result in at least XLT were selected (Figure 2A and supplemental Figure 3A).6-9 Similar to FL-WASp, all mutants translocate to the nucleus in TH1- and TCR-activation–dependent manner (reported by pTyr319 ZAP70) because their association with importin-β1 remains intact (supplemental Figure 3D-E).3 Because WASp-deficient TH cells demonstrate defect in TH0 > TH1 gene activation that is corrected by re-expressing FL-WASp,2,3 we asked whether re-expression of pathogenic WASp-mutants similarly restores this deficiency. Upon TH1-skewing, both WH1 mutants fail to restore the deficient expression of IFNG and TBX21 mRNA and protein, latter determined by ELISA and intracellular protein staining with flow cytometry (Figure 2B and supplemental Figure 4A-C). Dissimilarly, all non–WH1-domain mutants, including a PH-domain mutant, restore TH1-gene (mRNA and/or protein) expression to the level achieved with FL-WASp. Because these WH1 mutants clinically manifest XLT-to-WAS disease progression, whereas non–WH1 mutants (except PH-mutant S24F) do not,6-9 the observed mRNA and protein expression profiles of TH1 genes correlate with their respective disease severity grades. Notably, none of the mutants impaired TH2-driven upregulation of IL4 or GATA3 mRNA and protein (Figure 2B and supplemental Figure 4A-C), suggesting that the deleterious effect of WH1-domain mutations is context-specific.
Characterizing the effects of pathogenic WAS missense mutations on WASp:SWI/SNF associations in vivo. (A) Multidomain structure of WASp is shown along with the indicated pathogenic mutations within its different domains. Recurring “hot spot” mutations are indicated in red along with their reported clinical severity grades (stable XLT vs XLT>WAS 5A/5M progressive disease; 5A, grade 5 with autoimmunity; 5M, grade 5 with malignancy). See supplemental Figure 3A for a detailed description of the mutations and their corresponding disease severity grades. (B) RT-qPCR quantitation of candidate TH1 or TH2 genes in WASnull T-cell line (HTLV-1–immortalized) reconstituted with transfected FL-WASp or the indicated mutants after CD3/28 activation under TH1 or TH2 skewing or TH0 nonskewing conditions. Normal CD4 TH cell line (HTLV-1 immortalized) is the control. UT, untranfected WASnull T cells. The mRNA copy numbers derived from the control TH0 cells are not shown but were subtracted from the displayed final mRNA values of the TH1- or TH2-skewed cells. Absolute copy numbers adjusted to GAPDH are displayed as fold change (up or down) in TH1 or TH2 cells compared with their TH0 controls. Data represent the average from at least 3 biological replicates, with bars indicating SEM. Wilcoxon nonparametric test using GraphPad InStat software determined the P values comparing the data between FL and mutants (red asterisk, P < .01; black asterisk, P > .01 but ≤ .05). In data where the differences did not reach statistical significance (ie, P > .05), an asterisk is not shown. (C). MNase ChIP-qPCR assays were performed for the indicated proteins as described in the legend to Figure 1C. The genomic location of PCR primer/probes is indicated by a red asterisk in the gene diagram shown at the top. For TBX21, the 5′ UTR primers were designed within the genomic region that also contains a GAS (γ-activated sequence) site (5′-TTCAGGCAA-3′ at about −770 bp from first coding ATG). For IFNG, the primers are located between −200 to −250 bp from first coding ATG, a region known to contain functional promoter elements. The intergenic region between COL8A2 and TRAPPC3 genes on Chr.1, which does not contain known protein-coding genes, served as a negative control. See supplemental Table 1 for primer/probe details. (D). Nuclear fraction of Jurkat (TH1-skewed, TCR-activated) cells stably expressing Flag/Myc dual-tagged FL-WASp or its indicated WASp mutants were sequentially IPed in the indicated combination (eg, Flag>Myc denotes first IP: Flag, second IP: Myc) and analyzed by sequential western blotting of the same gel with the indicated antibodies.
Characterizing the effects of pathogenic WAS missense mutations on WASp:SWI/SNF associations in vivo. (A) Multidomain structure of WASp is shown along with the indicated pathogenic mutations within its different domains. Recurring “hot spot” mutations are indicated in red along with their reported clinical severity grades (stable XLT vs XLT>WAS 5A/5M progressive disease; 5A, grade 5 with autoimmunity; 5M, grade 5 with malignancy). See supplemental Figure 3A for a detailed description of the mutations and their corresponding disease severity grades. (B) RT-qPCR quantitation of candidate TH1 or TH2 genes in WASnull T-cell line (HTLV-1–immortalized) reconstituted with transfected FL-WASp or the indicated mutants after CD3/28 activation under TH1 or TH2 skewing or TH0 nonskewing conditions. Normal CD4 TH cell line (HTLV-1 immortalized) is the control. UT, untranfected WASnull T cells. The mRNA copy numbers derived from the control TH0 cells are not shown but were subtracted from the displayed final mRNA values of the TH1- or TH2-skewed cells. Absolute copy numbers adjusted to GAPDH are displayed as fold change (up or down) in TH1 or TH2 cells compared with their TH0 controls. Data represent the average from at least 3 biological replicates, with bars indicating SEM. Wilcoxon nonparametric test using GraphPad InStat software determined the P values comparing the data between FL and mutants (red asterisk, P < .01; black asterisk, P > .01 but ≤ .05). In data where the differences did not reach statistical significance (ie, P > .05), an asterisk is not shown. (C). MNase ChIP-qPCR assays were performed for the indicated proteins as described in the legend to Figure 1C. The genomic location of PCR primer/probes is indicated by a red asterisk in the gene diagram shown at the top. For TBX21, the 5′ UTR primers were designed within the genomic region that also contains a GAS (γ-activated sequence) site (5′-TTCAGGCAA-3′ at about −770 bp from first coding ATG). For IFNG, the primers are located between −200 to −250 bp from first coding ATG, a region known to contain functional promoter elements. The intergenic region between COL8A2 and TRAPPC3 genes on Chr.1, which does not contain known protein-coding genes, served as a negative control. See supplemental Table 1 for primer/probe details. (D). Nuclear fraction of Jurkat (TH1-skewed, TCR-activated) cells stably expressing Flag/Myc dual-tagged FL-WASp or its indicated WASp mutants were sequentially IPed in the indicated combination (eg, Flag>Myc denotes first IP: Flag, second IP: Myc) and analyzed by sequential western blotting of the same gel with the indicated antibodies.
WAS-causing but not XLT-causing mutations impair hBRM but not BRG1 promoter enrichment in vivo
To clarify the molecular basis for impaired TH1 but intact TH2 gene activation, we investigated chromatin-signaling events at TH1 and TH2 gene loci in WAS-null TH cells reconstituted with the representative WH1 mutants (T45M, R86C) or non–WH1 mutants (A236G, R477K, S24F). Despite intact nuclear localization of all mutants under TH1 skewing (supplemental Figure 3D-E), locus-specific recruitment of WH1 mutants (determined with both anti-WASp and anti-Flag antibodies in ChIP assays) at TBX21 and IFNG promoters is impaired compared with that of non–WH1 mutants (Figure 2C, first row; supplemental Figure 5C). Correspondingly, only WH1 mutants show a coordinate reduction in the promoter enrichment of BAF47 and BAF170, the 2 SWI/SNF subunits critical for optimal nucleosomal remodeling (Figure 2C, second row; supplemental Figure 5C).24 These WH1 mutants, however, do not impair promoter enrichment of BAF53 (ARP4) or β-actin, the 2 SWI/SNF subunits critical for BRG1 recruitment to chromatin and for its maximum adenosine triphosphatase (ATPase) activity (Figure 2C, third row).25 Consequently, recruitment of BRG1 to TH1-gene promoters is unaffected, whereas that of hBRM is impaired by WH1 mutants (Figure 2C, fourth row). WH1 mutants, however, do not disrupt promoter recruitment of hBRM/BAFs to CSF2 in TH1-skewed cells (Figure 2C) or to IL4 and GATA3 in TH2-skewed cells (supplemental Figure 4), implying that the deleterious effect of WH1 mutants is both gene- and TH1/TH2-differentiation–specific. In support of ChIP findings, sequential coIP (1stFlag>2ndMyc) on MNase-treated TH1-skewed cells also shows disrupted binding of WH1 mutants to BAF47, BAF170, and hBRM, whereas binding to BAF53, β-actin, or BRG1 is unaffected (Figure 2D). Because BRG1 and hBRM are present in separate CRCs classified as BAF (hBRM-containing) or PBAF (hBRM-devoid, BRG1-containing),16-18 our data propose a model wherein the deleterious effect of WH1 mutations on TH1 gene promoters is linked to impaired BAF but not PBAF chromatin-remodeling activity. A more direct evidence to support this model, however, will require RNAi-mediated depletion of BRG1 and hBRM in human T cells.
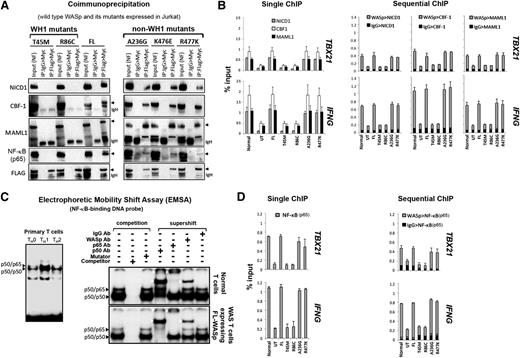
WAS-causing mutations that disrupt hBRM activity also impair Notch signaling
To determine whether WASp:hBRM association is functionally relevant in vivo, we tested the hypothesis that WASp effect on TH1-transcriptional reprogramming is linked with Notch, because hBRM but not BRG1 recruits Notch to gene promoters.26 Two-step coIP (1stFlag>2ndMyc) on MNase-treated TH1-skewed cells showed that the transfected FL-WASp binds endogenous Notch1-intracellular-domain (NICD1), CBF1/RBP-Jκ, and Mastermind-like1 (MAML1) (Figure 3A), a ternary complex that converts CBF1 from a repressor to an activator.27,28 Accordingly, single and sequential MNase-ChIP assays demonstrate FL-WASp coenriching with endogenous NICD1, CBF1, or MAML1 at the same promoter loci of IFNG or TBX21 (Figure 3B), which are both Notch-target genes.29-31 These data, suggestive of WASp:Notch alliance in TH1 activation, raised the possibility that WH1 mutations that impair BRM activity would contemporaneously impair Notch signaling at TH1-gene promoters.26 CoIP demonstrates that WH1 mutants (T45M, R86C) do not bind these Notch components, whereas non–WH1 mutants (A236G, R477K, S24F) bind them normally (Figure 3A and supplemental Figure 5B). Correspondingly, promoter coenrichment of Notch components is diminished in TH1-skewed cells expressing WH1 mutants, but not non–WH1 mutants, demonstrated by single and sequential ChIP assay (Figure 3B and supplemental Figure 5C). In TH2-skewed cells, however, chromatin enrichment of SWI/SNF- and Notch-complexes at IL4 and GATA3 promoters is unaffected in both WH1- and non-WH1 mutant–expressing cells (supplemental Figure 2B), implying context-dependent WASp specificity. We conclude that nuclear-WASp is required for both hBRM- and Notch-driven chromatin-signaling cascade(s) during TH1 but not TH2 differentiation, and that the “hot spot” missense mutations in the WH1 domain of WASp disrupt this signaling cascade.
Characterizing the effects of pathogenic WAS missense mutations on WASp:Notch:NF-κB signaling module in vivo. (A) CoIP/western assay. The same gel shown in Figure 2D was sequentially reprobed with the indicated Notch signaling components. (B,D) Single and sequential ChIP-qPCR assays were performed for the indicated proteins as described in the legend to Figure 1C. (C) Left panel: Electrophoretic mobility shift assay (EMSA) with NF-κB oligo performed on the nuclei isolated from primary human CD4+ TH cells, nonskewed TH0, or TH1- or TH2-skewed and CD3/28-activated cells. Right panel: EMSA/Supershift assay with NF-κB oligo performed on TH1-skewed normal TH cells (endogenous WASp) or WAS-null TH cells stably expressing Flag-tagged, transfected FL-WASp. The data are representative of 2 biological replicates. White arrowheads indicate location of the shifted bands.
Characterizing the effects of pathogenic WAS missense mutations on WASp:Notch:NF-κB signaling module in vivo. (A) CoIP/western assay. The same gel shown in Figure 2D was sequentially reprobed with the indicated Notch signaling components. (B,D) Single and sequential ChIP-qPCR assays were performed for the indicated proteins as described in the legend to Figure 1C. (C) Left panel: Electrophoretic mobility shift assay (EMSA) with NF-κB oligo performed on the nuclei isolated from primary human CD4+ TH cells, nonskewed TH0, or TH1- or TH2-skewed and CD3/28-activated cells. Right panel: EMSA/Supershift assay with NF-κB oligo performed on TH1-skewed normal TH cells (endogenous WASp) or WAS-null TH cells stably expressing Flag-tagged, transfected FL-WASp. The data are representative of 2 biological replicates. White arrowheads indicate location of the shifted bands.
WAS-causing, but not XLT-causing mutations, disrupt NF-κB (p65) activity at TH1-gene promoters
To further consolidate the WASp:BRM:Notch alliance in TH1 differentiation, we investigated the functional consequences of WH1 mutations on a downstream target of this signaling module. Because Notch1 and BRM both regulate NF-κB activity32,33 and both transcription factors (Notch, NF-κB) are critical in T-BET and IFN-γ production,29-31,34,35 we hypothesized that the deleterious effect of WH1 mutations on TH1 gene activation is linked to impaired NF-κB signaling. EMSA demonstrates increased DNA-binding activity of NF-κB in TH1-skewed compared with that in TH2-skewed or nonskewed human, primary TH0 cells (Figure 3C, left panel), which reinforces the role of NF-κB in TH1 differentiation.34,35 Significantly, EMSA/Supershift assays demonstrate that WASp is part of NF-κB:DNA ternary complex in TH1-skewed cells (Figure 3C).
WASp:NF-κB:DNA association in vitro reported by EMSA was validated in vivo by single and sequential ChIP. These demonstrate that WASp and NF-κB co-occupy the same promoter regions of IFNG and TBX21 in vivo in TH1-skewed cells (Figure 3D). Significantly, promoter enrichment of NF-κB(p65) is diminished only in WH1- but not non–WH1 mutant–expressing TH1-skewed cells (Figure 3D and supplemental Figure 5C). CoIP assays similarly show that only WH1- but not non–WH1 mutations disrupt binding of WASp to NF-κB(p65) (Figure 3A). The functional importance of WASp-Notch alliance, uncovered here for the first time in human cells, is critical also in Drosophila, where Wsp mutations result in impaired Notch-mediated cell-fate decisions.36 Together, our studies propose that nuclear-WASp impinges on chromatin-signaling cascade(s) that involves hBRM, Notch, and NF-κB during TH1-differentiation and identify Thr45 and Arg86 residues of WH1 domain of WASp to be important in this cascade.
WH1 mutations impair recruitment of histone H2A.Z and its catalyzing enzyme EP400 at TH1 gene promoters
We next investigated how the above chromatin-signaling defects affect chromatin remodeling. One mechanism to remodel chromatin is by exchanging histone H2A for H2A.Z, a process catalyzed by SWI2/SNF2-like proteins SRCAP or EP400 (hDomino).37-39 Because WASp binds EP400 in the nucleus as reported by MS and coIP (Figure 1A-B), and co-enriches with EP400 at the TH1-gene promoters in vivo as reported by ChIP (Figure 1C), we asked whether WH1 mutants disrupt EP400-dependent, H2A-to-H2A.Z exchange at WASp-target, TH1-gene promoters. CoIP shows that WASp binding to EP400 is disrupted by WH1- but not non–WH1 mutations (Figure 2D). Sequential ChIP shows impaired coenrichment of endogenous EP400 and transfected WH1 mutants at TH1-gene promoters (Figure 4A), which we show are also deficient in H3K4me3 modification in cells lacking nuclear-WASp2,3 or expressing WH1 mutants (Figure 4B). Because EP400 recruitment is positively regulated by H3K4me3,15 our finding raises the possibility that nuclear-WASp participates in H2A-to-H2A.Z exchange by regulating H3K4me3-dependent binding of EP400 to gene promoters. Indeed, WH1-mutants show a paucity of H2A.Z/H2B-marked heterotypic chromatin in the face of abundant unmodified H2A/H2B-marked chromatin at TH1-gene promoters, as reported by sequential ChIP (Figure 4A).
Characterizing the effects of pathogenic WAS missense mutations on histone H2A-to-H2A.Z exchange and transcription elongation in vivo. (A-B) Single and sequential MNase-ChIP assays performed on nuclear extracts from TH1-skewed, WASnull TH cells stably expressing the indicated WASp mutants. Other details are as described in the legend to Figures 2C and 3B. Data represent the averages of triplicates, with bars indicating the standard error from at least 3 biological replicates. (C) PCR-based quantitation of the absolute DNaseI hypersensitivity at the indicated gene promoter loci in nonskewed TH0 or TH1-skewed, normal, and WASnull TH cells stably expressing the indicated WASp mutants, or controls (UT, untransfected control; FL, full-length WASp transfected) is shown. TH nuclei were treated with DNaseI, and the number of amplicons lost was quantified by RT-qPCR and is displayed as percent of DNaseI-untreated control. Data represent the average of triplicate values from 1 experiment. (D) MNase-ChIP assays performed on nuclear extracts from TH1-skewed, WASnull TH cells stably expressing the indicated WASp mutants. The location of 8 different genomic positions (position 1 is near TSS at 5′UTR; position 8 is at 3′UTR immediately after the last coding exon) within the IFNG locus, where the ChIP-qPCR primer/probes were designed is indicated and numbered (in red). DNaseI HS (DHS) profile for the primary human peripheral TH1 cells (in gray) publicly available from the ENCODE–University of Washington was aligned alongside our custom track to give context to the location of our ChIP-qPCR primer/probes. “Normal” denotes an HTLV-1–immortalized CD4+T cell line generated from a healthy donor. UT, untransfected WAS-null patient-derived T cells line.
Characterizing the effects of pathogenic WAS missense mutations on histone H2A-to-H2A.Z exchange and transcription elongation in vivo. (A-B) Single and sequential MNase-ChIP assays performed on nuclear extracts from TH1-skewed, WASnull TH cells stably expressing the indicated WASp mutants. Other details are as described in the legend to Figures 2C and 3B. Data represent the averages of triplicates, with bars indicating the standard error from at least 3 biological replicates. (C) PCR-based quantitation of the absolute DNaseI hypersensitivity at the indicated gene promoter loci in nonskewed TH0 or TH1-skewed, normal, and WASnull TH cells stably expressing the indicated WASp mutants, or controls (UT, untransfected control; FL, full-length WASp transfected) is shown. TH nuclei were treated with DNaseI, and the number of amplicons lost was quantified by RT-qPCR and is displayed as percent of DNaseI-untreated control. Data represent the average of triplicate values from 1 experiment. (D) MNase-ChIP assays performed on nuclear extracts from TH1-skewed, WASnull TH cells stably expressing the indicated WASp mutants. The location of 8 different genomic positions (position 1 is near TSS at 5′UTR; position 8 is at 3′UTR immediately after the last coding exon) within the IFNG locus, where the ChIP-qPCR primer/probes were designed is indicated and numbered (in red). DNaseI HS (DHS) profile for the primary human peripheral TH1 cells (in gray) publicly available from the ENCODE–University of Washington was aligned alongside our custom track to give context to the location of our ChIP-qPCR primer/probes. “Normal” denotes an HTLV-1–immortalized CD4+T cell line generated from a healthy donor. UT, untransfected WAS-null patient-derived T cells line.
WH1 mutations disallow development of increased DHS at TH1 gene promoters
Because H2A/H2B-marked hybrid nucleosomes do not reposition as well as H2A.Z/H2B-marked nucleosomes,40 we performed qPCR-based DHS assay to determine the degree of promoter compaction. In cells expressing normal or FL-WASp or non–WH1 mutants, TH1 activation results in increased hypersensitivity of the TH1-gene promoters reported in fewer amplicons remaining after DNase I treatment. By contrast, in the WH1 mutants, the number of amplicons in TH0 and TH1 cells remains essentially unchanged, suggesting suboptimal promoter decompaction of IFNG and TBX21 promoters (Figure 4C). This DHS defect is not observed at the CSF2 locus, a WASp–non-target gene in TH1 cells. Moreover, we find that decreased enrichment of hBRM at gene promoters in WH1 mutant–expressing TH1 cells occurs contemporaneously with increased enrichment of EZH2 and the repressive histone mark H3K27me3 (Figure 4B). Because EZH2-like proteins inhibit recruitment and function of SWI/SNF at the promoters,41 we propose that this defect might further add to the decreased accessibility of nucleosomal DNA at gene promoters in WH1 mutant–expressing cells.
WH1 mutations impede transcription elongation of IFNG beyond first intron
To determine how H2A.Z-related, chromatin-remodeling defect affects transcription along the DNA translational axis, we investigated transcription initiation and elongation steps at the IFNG locus in TH1 cells. We mapped ChIP enrichment profiles of RNA Pol II pCTD-Ser5 (transcription-initiation mark), pCTD-Ser2, and CDK9 (both transcription-elongation marks) at 8 different sites within the IFNG locus spanning 5′UTR to 3′UTR (Chr.12: 68 548 000- 68 554 000) (Figure 4D). In TH1 cells expressing normal WASp, enrichment of pCTD-Ser5 is highest between 5′-TSS and exon-1/intron-1 genomic region, whereas that of pCTD-Ser2 and CDK9 (P-TEFb subunit) are highest toward 3′-end, with the crossover occurring at ∼500 nt downstream of TSS. Such a CTD code identifies most actively transcribed genes.42 By contrast, in WH1 mutant–expressing TH1 cells, enrichments of pCTD-Ser2 and CDK9 are diminished at the mid–intron-1 region, this despite normal enrichment of pCTD-Ser5 at 5′UTR (Figure 4D). Accordingly, our results endorse the paradigm that H2A.Z-enriched nucleosomes optimize transcription elongation by relieving stalled Pol II within the gene body in a role that is linked to pCTD-Ser2.43,44 Moreover, because both hBRM and NF-κB(p65) regulate transcription elongation,45,46 we propose that hBRM and p65 defects also contribute to abnormal TH1-transcriptional reprograming in a subset of WAS patients carrying hot-spot WH1 mutations.
Discussion
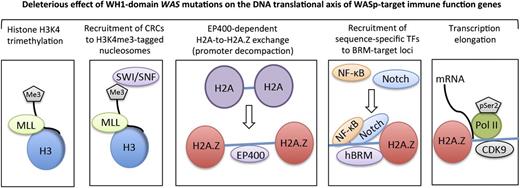
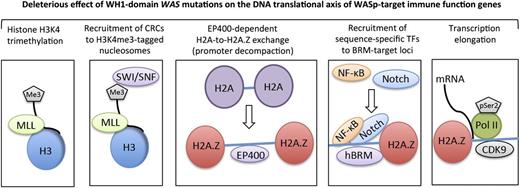
The clinical phenotypes of genetic disorders are influenced by epigenetic changes that affect gene expression.47-50 Although epigenetics provide a powerful way to stratify risk genotypes in diseases, how a disease-causing point mutation influences the epigenetic code of the target cell type in which the gene product is expressed is not well understood. Using a disease model of WAS, we provide a SWI/SNF-linked mechanism by which a subset of WAS missense mutations differentially perturb transcription of “core” genes that pattern TH1-adaptive immunity. We show that a subset of SWI/SNF complexes require functional WASp to execute their locus-specific, chromatin-remodeling functions during TH1 differentiation in human T cells. Most importantly, the 2 hot-spot WAS missense mutations that clinically manifest XLT-to-WAS disease progression disrupt SWI/SNF activity, whereas mutations causing stable XLT do not. This suggests a direct relationship between gene mutation and epigenetic disease–driving events within the context of TH1 differentiation for the pathogenic mutations tested. Several lines of evidence propose nuclear-WASp to be an important component in the chromatin-remodeling process during TH1 development: (1) Nuclear-WASp binds multiple CRCs, in vivo, as determined by MS and coIP; (2) nuclear-WASp co-ChIPs with these CRCs at the same cis-regulatory genomic sites in TH cells; and (3) WH1 domain, WAS missense mutations disrupt recruitment of CRCs with deleterious consequences on histone H2A-to-H2A.Z exchange at WASp-target gene promoters. Therefore, compared with cytosolic WASp, which participates in cytoskeletal remodeling via ARP2/3-complexes, nuclear-WASp participates in chromatin remodeling via hSWI/SNF-complexes (Figure 5).
A working model for nuclear-WASp actions in reprogramming transcription. Step 1: H3K4me3 trimethylation. Because nuclear-WASp physically and functionally associates with the hMLL/COMPASS complex and histone H3K4 trimethylation,2,3 the model proposes that WH1 mutants disrupt recruitment of MLL-enriched complex and the subsequent inscription of H3K4me3 mark at WASp-target gene promoters. Step 2: Recruitment of CRCs to H3K4me3-tagged nucleosome. Because the human SWR1-like protein EP400 favors binding promoters that are enriched with H3K4me3-marked nucleosomes,15 the model proposes that WASp regulates EP400 binding to chromatin in TH1 cells through its previously described effect on H3K4me3 modification.2 WH1 mutations disrupt this function of WASp at its target loci. Step 3: EP400-dependent H2A-to-H2A.Z exchange. Recruitment of EP400 to H3K4me3-marked chromatin catalyzes local H2A-to-H2A.Z exchange, which promotes promoter decompaction. Because the augmented recruitment of SWI/SNF occurs at genomic sites containing H2A.Z-tagged nucleosomes,39 the model proposes that WH1 mutants disrupt the initial H2A.Z-driven chromatin-remodeling and consequently also the deposition of SWI/SNF-like BAF subunits, which in turn affects higher-order chromatin reorganization and RNA polymerase II recruitment.18,24 Step 4: Recruitment of sequence-specific transcription factors (TFs). WH1 mutants disrupt recruitment of Notch-signal transduction components and NF-κB(p65) at the hBRM target loci in TH1 cells. Step 5: Transcription elongation. EP400-dependent H2A.Z deposition functions also relieve the RNA Pol II “pause” at +1 nucleosome.43,44 We propose that WH1 mutants, through their effects on EP400 and H2A-to-H2A.Z exchange, disrupt recruitment of elongating Pol II (CTD-Ser2) and CDK9 (PTEF-b subunit), protein complexes that actuate productive 5′>3′ transcription elongation. The disease model of WAS/XLT proposes that certain WAS mutations could impair one or multiple steps in this processive event, leading to a spectrum of defects that could manifest in either total loss of gene transcription or some gradations of it, which we postulate lends the immunologic basis for cross-phenotype effects and symptom heterogeneity in XLT/WAS.
A working model for nuclear-WASp actions in reprogramming transcription. Step 1: H3K4me3 trimethylation. Because nuclear-WASp physically and functionally associates with the hMLL/COMPASS complex and histone H3K4 trimethylation,2,3 the model proposes that WH1 mutants disrupt recruitment of MLL-enriched complex and the subsequent inscription of H3K4me3 mark at WASp-target gene promoters. Step 2: Recruitment of CRCs to H3K4me3-tagged nucleosome. Because the human SWR1-like protein EP400 favors binding promoters that are enriched with H3K4me3-marked nucleosomes,15 the model proposes that WASp regulates EP400 binding to chromatin in TH1 cells through its previously described effect on H3K4me3 modification.2 WH1 mutations disrupt this function of WASp at its target loci. Step 3: EP400-dependent H2A-to-H2A.Z exchange. Recruitment of EP400 to H3K4me3-marked chromatin catalyzes local H2A-to-H2A.Z exchange, which promotes promoter decompaction. Because the augmented recruitment of SWI/SNF occurs at genomic sites containing H2A.Z-tagged nucleosomes,39 the model proposes that WH1 mutants disrupt the initial H2A.Z-driven chromatin-remodeling and consequently also the deposition of SWI/SNF-like BAF subunits, which in turn affects higher-order chromatin reorganization and RNA polymerase II recruitment.18,24 Step 4: Recruitment of sequence-specific transcription factors (TFs). WH1 mutants disrupt recruitment of Notch-signal transduction components and NF-κB(p65) at the hBRM target loci in TH1 cells. Step 5: Transcription elongation. EP400-dependent H2A.Z deposition functions also relieve the RNA Pol II “pause” at +1 nucleosome.43,44 We propose that WH1 mutants, through their effects on EP400 and H2A-to-H2A.Z exchange, disrupt recruitment of elongating Pol II (CTD-Ser2) and CDK9 (PTEF-b subunit), protein complexes that actuate productive 5′>3′ transcription elongation. The disease model of WAS/XLT proposes that certain WAS mutations could impair one or multiple steps in this processive event, leading to a spectrum of defects that could manifest in either total loss of gene transcription or some gradations of it, which we postulate lends the immunologic basis for cross-phenotype effects and symptom heterogeneity in XLT/WAS.
Interestingly, not all WAS mutations are damaging to SWI/SNF activity, a finding that correlates with their respective clinical severity grades. Accordingly, WH1 mutations (T45M, R86C) that manifest XLT-to-WAS progression disrupt binding of mutant WASp to SWI/SNF complexes and its recruitment to TH1-gene promoters, whereas mutations of the basic-domain (A236G) or VCA-domain (R477K) that manifests in XLT do not disrupt these molecular readouts. Inexplicably, whereas 2 of 3 S24F (PH-domain mutation) reported patients manifest XLT and thus conform to the “molecular signatures” identified for other non-WH1 mutants, 1 patient manifests CP effects (XLT-to-WAS progression). Similarly, whereas 3 D485G (VCA mutation) reported patients manifest XLT, 1 patient (D485N) manifests CP effects. Because D485N does not impair ARP2/3-driven actin-nucleation,51 whether this VCA-domain mutation has a “long-range” effect of disrupting SWI/SNF activity is a possibility that awaits experimental validation.
Notably, the deleterious effect of WH1 mutations on SWI/SNF is limited to certain subunits, being most pronounced on BAF170 and BAF47 but not on BAF53 (ARP4) or β-actin. Because incorporation of BAF170 and BAF47 into nascent-SWI/SNF complex is required for optimal nucleosomal remodeling,23 our findings suggest that WH1 mutations disrupt neoassembly of higher-order CRCs at WASp-target gene promoters. This BAF defect occurs coordinately with an increased occupancy of EZH2 methylase (polycomb-group family) and the consequent enrichment of histone H3K27me3, a mark of “transcriptionally silent” chromatin. This antagonism between CRCs and EZH2, which has been demonstrated for BAF47 and EZH2 in tumorigenesis,52 might further contribute toward suppressing TH1-gene activation.
Another interesting finding is that the 2 hot-spot WH1 mutations selectively impair promoter recruitment of hBRM but not BRG1, the 2 homologous central ATPases of mammalian SWI/SNF complexes that direct distinct cellular processes. BRM targets Notch-regulated gene promoters, whereas BRG1 targets gene promoters regulated by zinc-finger proteins.26 Consequently, we show that promoters lacking hBRM contemporaneously lack recruitment of the core components of Notch signaling in vivo, resulting in impaired chromatin-activity of NF-κB, a downstream Notch1 target.32 This finding is immediately relevant to the immune dysregulation in WAS, because NF-κB is critical for TH1 development.35 Because removal of H2A/H2B-containing nucleosomes is required for the specific binding of NF-κB,53 we propose that WASp through its effects on H2A-to-H2A.Z exchange facilitates NF-κB binding to gene promoters in TH1 cells. Therefore, BRG1, whose enrichment at TH1-gene promoters is unaffected, cannot substitute for the lost hBRM functions during TH1 transcriptional-reprogramming. This suggests that the functional effects of BRG1 and BRM are influenced by the lineage-specific transcription factors they coassociate with, a paradigm that supports the “Activator Model” for SWI/SNF recruitment to promoters.54 However, we cannot rule out the possibility that the promoter-enriched BRG1 maybe “nonfunctional,” and, if so, BRG1 defect could also contribute to impaired chromatin signaling.
In addition to its effects on hBRM/BAF promoter activity, WH1 mutations perturb other reaction steps of the multistep chromatin-remodeling process.55,56 WH1 mutants interfere with the enrichment of H2A/H2A.Z-marked heterotypic chromatin at TH1-gene promoters via its effects on EP400, hSWR1/hDomino-homolog that catalyzes H2A.Z deposition in vivo.20,39 Because H2A.Z imparts increased mobility to the peri-TSS nucleosomes,57 the H2A.Z deficiency observed with WH1-domain mutants affects chromatin decompaction at TH1-gene promoters reported in a DHS profile that is nonconducive to recruiting transcription factors.58 Because H2A.Z is also important in DNA repair, determining whether and how WASp effects on H2A-to-H2A.Z exchange at other genomic loci affects this process will begin to clarify why certain WAS mutations associate with genomic instability in lymphocytes.59 Given the importance of chromatin remodeling in biology, including in cancer development,48,49,60-63 we predict that the WASp:SWI/SNF alliance may affect multiple other-cell biological processes including tumor suppression, the disruption of which may further influence the phenotypic variations in WAS. Notwithstanding, the implications of our studies should be limited to the TH1 development, because we have not studied megakaryocytes, the major cell type responsible for the early findings of thrombocytopenia (XLT) in WAS. Although thrombocytopenia was shown to be VCA-domain–independent,64 clarification of whether WAS mutations differ in their proclivity to perturb other aspects of megakaryocyte biology is essential. Irrespective of the multiple potential pathways that will eventually be determined to contribute to the complete disease-causing spectrum of WAS, our study provides the beginnings of a novel epigenetic basis to predict symptom severity in WAS, at least as contributed by TH cell dysregulation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Jerome Parness (University of Pittsburgh) for insightful discussions and critical reading of the manuscript, M. Balasubramani of the University of Pittsburgh Proteomic Core Facility for analyzing the MS data, and Fabio Candotti for providing WAS patient T-cell line. We thank the University of Iowa Dance Marathon for providing laboratory space.
This research was supported by National Institutes of Health National Institute of Allergy and Infectious Diseases grants R01 AI073561 and R01 AI084957.
Authorship
Contribution: K.S. performed the majority of the experiments; S.S. generated all patient mutants and assisted in ChIP and PCR assays; S.-S.H. performed the EMSA/Gel-shift assays; and Y.M.V. conceived the study, designed the experiments, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yatin M. Vyas, Division of Pediatric Hematology-Oncology, University of Iowa Children’s Hospital, Iowa City, IA 52242; e-mail: yatin-vyas@uiowa.edu.