Abstract
Introduction: The PI3K pathway plays a significant role in cell cycle, apoptosis, and DNA repair. It is commonly dysregulated in cancers making it a potential therapeutic target. BKM120 is a novel oral pan-class I PI3K inhibitor with antitumor activity and efficacy reported in solid tumors. It is in phase I/II clinical trials for treatment of relapsed-refractory NHL. There are additional data desired towards molecular mechanism of action and biological pathways of resistance for BKM120-treated lymphoma.
Methods: TCL cell lines (Jurkat, Hut78, HH), HL lines (L428, L540) and DLBCL lines (SUDHL4, SUDHL6, SUDHL10, OCILY3, OCILY19) were treated with increasing concentrations of BKM120 (0.16-10uM) in 96 well plate and cell viability assessed by MTT assay. For gene expression profiling (GEP), SUDHL6, OCILY3, Jurkat, Hut78 and L540 cells were treated at IC50and analyzed on Illumina human HT12 gene chip. Gene Set Enrichment Analysis (GSEA) and biological network analysis were done using Ingenuity Pathway Analysis and Cytoscape. Apoptosis was evaluated by Annexin V and propidium iodine (AV/PI) staining and flow cytometry. For cell cycle analysis, Jurkat cells were exposed to BrdU and stained with anti-BrdU FITC and 7-AAD and analyzed by flow cytometry.
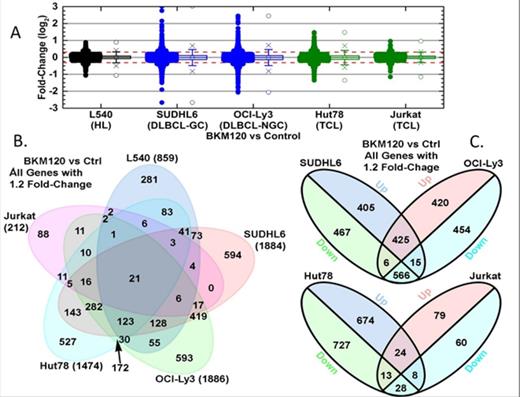
Results: Cell viability showed dose-dependent growth inhibition in all lines. IC50 was between 0.316μM - 3.72μM at 72 hours (hrs). GEP analyses showed significant gene changes following BKM120 treatment with a 1.2 fold-change for OCILY3 (1886 genes), SUDHL6 (1884 genes), Hut78 (1474 genes), L540 (859 genes) and Jurkat (212 genes) (Fig 1). Differential gene expression with BKM120-treated cells showed significant overlap among DLBCL cell lines, with 991 genes of 1886 (OCILY3) and 1884 (SUDHL6). The differentially expressed genes in L540 HL cells treated with BKM120 showed better overlap with DLBCL lines than with TCL lines, with 366 and 315 of 859 L540 genes overlapping with OCILY3 and SUDHL6 lines, respectively. This may reflect the B cell of origin related response to BKM120 treatment. TCL lines had different profiles, with 52 overlapping genes of 212 for Jurkat and 1474 for Hut78. Jurkat is known to harbor a PTEN mutation with constitutively active PI3K, which may explain the differences in the GEP observed with BKM120 treatment in Jurkat and Hut78 TCL. GSEA and biological network analysis of BKM120 treated lymphoma cells showed conserved inhibitory effects on cell cycle, DNA replication and metabolic process across all lymphoma cells. However, we observed that immune signaling processes were oppositely regulated in B and T cell lymphomas. AV/PI-staining and flow cytometry revealed dose-dependent increase of apoptotic cells in all lymphoma cell lines. Analysis based on BrdU incorporation revealed induction of G2/M arrest in Jurkat cells treated with BKM120. Western blots showed decreased phosphorylation of PI3K and mTOR substrates including 4-EBP and phospho-p70S6K with BKM120 treatment. Although phosphorylation of MEK/ERK were downregulated at lower doses of BKM120, at higher doses, increased MEK/ERK phosphorylation were seen. Analysis of cell cycle regulatory proteins Cyclin, CDK and phospho-histone H3 with BKM120 treatment resulted in expression changes consistent with G2/M arrest and also resulted in increased apoptosis related PARP cleavage in all lymphoma cell lines.
Conclusions: BKM120 treatment elicited biologically distinct GEP with immune functions oppositely regulated in B and T cell lymphomas, however impairment of cell cycle, DNA replication and metabolic function were the core responses/function to PI3K inhibition. Furthermore, based on GSEA, it was predicted that cell cycle arrest and MAPK activation as likely mechanisms of resistance to BKM120 treatment in these lymphoma cell lines. Additional studies are planned to examine rational combinations of BKM120 together with other targeted small molecules (e.g., Chk1-inhibitor, CDK4/6 inhibitor and MEK inhibitor).
Differential gene expression and analysis of gene overlap. A. Point and boxplot representations of changes in gene expression following treatment with BKM120 for each cell line. B. Venn diagram of genes changing by 1.2 fold or more following treatment with BKM120 by cell line. C. Venn diagram of genes changing by 1.2 fold or more following treatment with BKM120, comparing SUDHL6 (Germinal center DLBCL) and OCI-Ly3 (ABC-DLBCL) and Hut78 and Jurkat (TCL lines).
Differential gene expression and analysis of gene overlap. A. Point and boxplot representations of changes in gene expression following treatment with BKM120 for each cell line. B. Venn diagram of genes changing by 1.2 fold or more following treatment with BKM120 by cell line. C. Venn diagram of genes changing by 1.2 fold or more following treatment with BKM120, comparing SUDHL6 (Germinal center DLBCL) and OCI-Ly3 (ABC-DLBCL) and Hut78 and Jurkat (TCL lines).
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.