Abstract
Background: Chronic lymphocytic leukemia (CLL) is considered as typical malignancy with ethnic difference, which is one of the common leukemia in western countries, while extremely rare in Asian countries. In addition to incidence rates, mean age of CLL in Asian is younger than that of Caucasian. Recently, many report using next generation sequencing revealed predictive mutations with prognostic implication and treatment response in Caucasian. Yet, genetic profiles of CLL and its prognostic significance have rarely been reported in Asian. To investigate whether genomic profile of CLL in Korea and prognostic impact of adverse mutation shows ethnic difference from Caucasian, we performed target capture sequencing of 87 hematologic malignancy related genes and fluorescent in situ hybridization for prognostic cytogenetic aberrations.
Methods: A total of 71 CLL patients were enrolled (median age at diagnosis 61 years, range 23-81 years). Conventional cytogenetic study (n=60) and FISH for chromosome 12 enumeration, 13q14.3 deletions, and 17p13 deletions (n=51) were performed. Target exome sequencing for an 87-gene panel was performed using IlluminaHiSeq 2500 (n=48). Mutations were selected by running algorithms including SIFT, Polyphen2, and CADD, and normal variant was filtered by by 1000Gp, ESP6500, and the in-house Korean database (n=250). Candidate mutations were confirmed by Sanger sequencing. We assessed disease progression, survival and response to treatment.
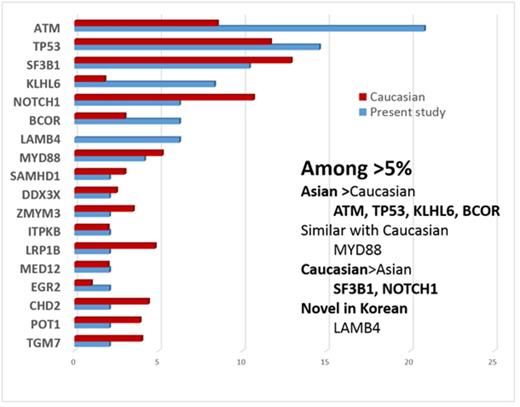
Results: Overall, 37 of 48 patients (77.1%) had at least one mutation; the mean number of mutations per patient was 2.1 (range 0-6). The average coverage of target regions was 200-fold. Targeted exome-sequencing analysis revealed 78 substitutions and insertion/deletions. The most common recurrent mutation (>5% frequency) was ATM (20.8%), followed by TP53 (14.6%), SF3B1 (10.4%), KLHL6 (8.3%), LAMB4 (6.25%), BCOR (6.25%) and NOTCH1 (6.25%). Compared to Caucasian, the ATM, KLHL6 and BCOR mutationsshowed higher frequency in Korean, while NOTCH1, LRP1B, CHD2, POT1 and TGM7 mutations were more frequent in Caucasian. Of note, the ATM mutation exhibited a 2-fold higher incidence (20.8% vs. 9%) in Koreans compared to Caucasians. Frequencies of TP53, SF3B1, MYD88, FAT4, SAMHD, DDX3X, ITPKB, and MED12 mutations were similar between Korean and Caucasian. The LAMB4, SH2B3, RUNX1, SCRIB, KIT, GATA2, CEBPA, TCF12, STAG2, ZRSR2, SETBP1, CSF1R, and SETBP1 mutations have not been reported in Caucasians. Mutations with adverse prognostic implication in Korean CLL was different from those of Caucasian. The TP53, MYD88, SF3B1, ITPKB, SAMHD1, and EGR2 mutations were associated with poor prognosis (P =0.023, P =0.005, P =0.032, P =0.011, P =0.049 and P =0.032, respectively, conventional Cox regression model, P <0.05). Aberrant karyotypes were observed in 28.3% (17/60) of patients by G-banding and in 66.7% (34/51) of patients by FISH. Compared to the reports of western countries, frequency of cytogenetic aberrations by FISH is similar and 13q14 deletion (45.8%, 22/48) was the most frequent chromosomal aberration, follow by trisomy 12 (30.0%, 15/50) and 17p deletion (23.5%, 12/51).
Conclusions: The somatic mutation profiles and adverse prognostic genes of Korean CLL differed from that of Caucasian CLL patients, while cytogenetic profiles were similar. Novel mutations discovered in this study must be validated through large cohort studies and may offer clues to the mechanisms underlying the ethnic difference in CLL incidence. In the future, therapeutic strategies targeting these genes should be evaluated and considered. To our knowledge, this is the first comprehensive mutation analysis of CLL patients in Asians using next generation sequencing.
Comparison of mutation frequencies (%) between Caucasian and Korean populations
Comparison of mutation frequencies (%) between Caucasian and Korean populations
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.