Abstract
Introduction: Focal lesions (FL) are detected by magnetic resonance imaging (MRI) and positron emission tomography (PET) and precede the development of osteolytic lesions in multiple myeloma (MM). FLs are absent in most patients with benign disease and their detection is associated with earlier disease progression suggesting that a distinct MM cell niche in the FLs is associated with conditions that promote the transition to MM. Studying the nature of this niche can significantly enhance our understanding of the biology and progression of MM.
Methods: Random BM aspiration samples were taken from the posterior superior iliac crest whereas FL samples were sampled under CT guidance from newly diagnosed MM patients. Gene expression profiling (GEP) was performed on CD138-enriched plasma cells (PC, n=170) and non-enriched BM trephine biopsies (n=49) from paired RBM and FL samples from the same patients and from unrelated RBM cases with no detectable FL (n=79). 8-multicolor flow cytometry (MFC) analysis was performed on 25 paired PC samples and selected genes were validated using immunohistochemistry (IHC).
Results: AComparison of GEP from paired RBM-PC and FL-PC showed discrepancies in GEP-based risk score and molecular subgroups and lower Polyclonal-PC score (reflecting the proportion of normal PC infiltration) in FL-PC samples (p=0.0001). There is 89% concordance for the GEP70 risk signature with 10 patients having low-risk PCs in RBM but high-risk PCs in the paired FL, and 8 patients showing high-risk PCs in RBM but low-risk PCs in the paired FL samples. In this setting progression-free and overall survival are mediated by the presence of a high-risk score in either sample. When molecular subgroups were identified there were more high risk-associated PR cases in FL samples (n=28) compared to only 16 PR cases in respective paired RBM-PC samples (p=0.005).
Flow cytometry data from 25 paired MM cell samples showed consistently lower surface expression of CD138 in FL (p=0.0001). Cases with detectable CD81 had lower CD81 expression on FL-PC (p=0.03). Discrepancies were also observed in cell surface expression of CD38 and CD45.
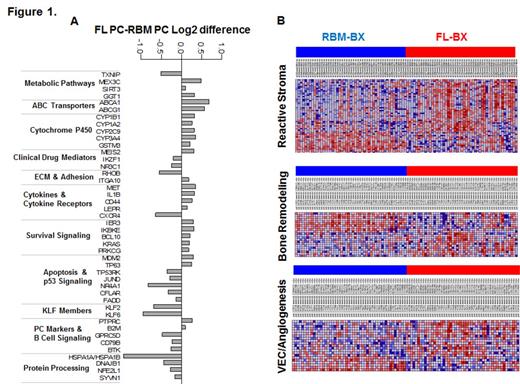
Pathway's analysis was based on of 523 differentially expressed genes between paired FL and RBM-PC samples (n=170; FDR<0.001) after adjusting for the level of normal PC infiltration. The top KEGG-based pathways enrichment analysis were associated with energy and drug metabolism, survival, cell-cell contact interaction and factors involved in activity of dexamethasone (e.g. NR3C1) and IMiDs (e.g. IZKF1), Figure 1A. Differential expression of ABCA1, the most upregulated gene in FL, was validated by IHC in biopsies. To test whether PCs from patients with FL have specific characteristics irrespective of their location, we compared RBM-PC GEP with RBM-PC GEP of unrelated patients with no detectable FLs. We detected increased expression of cell cycle genes in PC from patients with detectable FL.
Reduced osteogenesis and interaction with mesenchymal and vascular lineages have been linked with MM cell phenotypes and dissemination in BM. Microenvironmental reactive stroma (e.g. POSTN, collagen genes) and angiogenic (e.g. EDNRA) gene signatures were significantly upregulated in non-enriched FL biopsies albeit expected high proportion of PC in this site, whereas osteoblastic markers such as BGLAP and IBSP were underexpressed in these samples in comparison to paired non-enriched RBM trephine biopsies, Figure 1B.
Conclusions: PC in FL and RBM sites of the same patient are heterogeneous in their phenotype, molecular classification based on risk-score and subgroups, and pathways. GEP signatures of MM cells and the stroma in this niches stressing the biological and clinical relevance of FL as a hallmark in MM.
Yaccoby:University of Arkansas for Medical Sciences: Employment. Epstein:University of Arkansas for Medical Sciences: Employment. Johnson:University of Arkansas for Medical Sciences: Employment. Qu:Cancer Research and Biostatistics: Employment. van Rhee:University of Arkansa for Medical Sciences: Employment. Jethava:University of Arkansas for Medical Sciences: Employment. Stein:University of Arkansas for Medical Sciences: Employment. Mitchell:Cancer Research and Biostatistics: Employment. Heuck:Millenium: Other: Advisory Board; University of Arkansas for Medical Sciences: Employment; Celgene: Consultancy; Janssen: Other: Advisory Board; Foundation Medicine: Honoraria. Davies:Millenium: Consultancy; Janssen: Consultancy; Celgene: Consultancy; University of Arkansas for Medical Sciences: Employment; Onyx: Consultancy. Crowley:Cancer Research and Biostatistics: Employment. Weinhold:Janssen Cilag: Other: Advisory Board; University of Arkansas for Medical Sciences: Employment. Barlogie:University of Arkansas for Medical Sciences: Employment. Morgan:Takeda: Honoraria, Membership on an entity's Board of Directors or advisory committees; Bristol Myers Squibb: Honoraria, Membership on an entity's Board of Directors or advisory committees; CancerNet: Honoraria; Weismann Institute: Honoraria; MMRF: Honoraria; University of Arkansas for Medical Sciences: Employment; Celgene: Honoraria, Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.