Abstract
Epigenetic modulators have emerged as promising targets for treating cancers, especially blood cancers. As the major histone H3K4 methylation enzymes in mammals, the SET1/MLL complexes represent potential drug targets in epigenetic therapeutics due to (i) the intimate connection of H3K4 methylation with gene expression, and (ii) their extensive association with multiple cancers including blood cancers. However, the functional role for the SET1/MLL complexes in tumorigenesis remains largely unclear.
The SET1/MLL complexes comprise one of six different catalytic subunits and several shared core subunits including DPY30. We have previously shown that DPY30 directly facilitates genome-wide H3K4 methylation, and plays a crucial role in fundamental cellular processes including proliferation and differentiation, especially in the hematopoietic system. Our new analyses have shown that the core, but not the catalytic, subunits of SET1/MLL complexes is significantly up-regulated in primary human Burkitt's lymphomas bearing MYC-Ig translocations compared to other B lymphomas, and Myc binds to genes encoding the core but not the catalytic subunits. These results indicate that the core subunits are directly regulated by MYC, and prompted us to study their functional role in MYC-driven tumorigenesis.
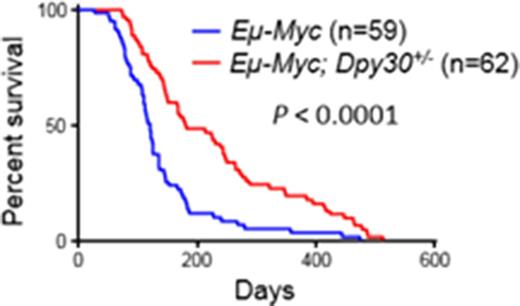
Using a Dpy30 conditional knockout mouse model that we recently established, we have shown a critical role of Dpy30 in the fate determination of hematopoietic stem and progenitor cells. Due to the severe pancytopenia of the knockout mice, we tested if genetically reducing Dpy30 dose may affect Myc-driven tumorigenesis in the Eμ-myc mouse. We found that Eμ-myc; Dpy30+/- mice survived significantly longer than their Eμ-myc littermates (see figure), with the median survival extended from 121 to 180 days, and with significantly alleviated spleen enlargement. Importantly, Dpy30+/- mice (no Eμ-myc) appear completely healthy with normal blood profiles. These results demonstrate that reducing Dpy30 level confers a significant resistance to Myc-driven lymphomagenesis without affecting normal physiology. We then found that, in the presence of Eμ -Myc, Dpy30 heterozygosity significantly increased apoptosis of splenic B cells, and reduced expression of some key anti-apoptotic genes. We further showed that Dpy30 directly bound to and controlled the H3K4 methylation at the regulated anti-apoptosis genes in splenic B cells. These results suggest that Myc overexpression increases the dependence of key apoptosis-regulatory genes on Dpy30, and thus sensitizes tumor cells to Dpy30 inhibition, exhibiting "epigenetic vulnerability".
To further study DPY30's role in MYC-dependent tumorigenesis at the molecular level, we have shown that DPY30 depletion in a MYC-dependent B lymphoma cell line markedly reduced (i) the lymphoma cell growth, (ii) expression of MYC targets, and most interestingly, (iii) binding of MYC to many of its genomic targets, as revealed by our ChIP-seq results. These results suggest that, in addition to promoting the expression of MYC gene itself that we previously found, DPY30 also reguates MYC's activity through promoting the genomic binding of MYC protein for target transcription.
Taken together, our studies have established an important role of Dpy30 in the Myc-driven lymphomagenesis, partially through its regulation of the target binding activity of Myc. Further studies of the genome-wide impact of Dpy30 inhibition on the chromatin configuration and expression of key tumoregenic genes are undergoing and will be discussed. These studies will help us understand how Dpy30-mediated chromatin modification coordinates with key oncogenes in promoting hematological malignancies, and thus may represent a potential epigenetic target in treatment of certain blood cancers.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.