Abstract
Aneuploidy causes a proliferative disadvantage, mitotic and proteotoxic stress in non-malignant cells ( Torres et al. Science 2007). Chromosome gain or loss, which is the hallmark of aneuploidy, is a relatively common event in Acute Myeloid Leukemia (AML). About 10% of adult AML display isolated trisomy 8, 11, 13, 21 (Farag et al. IJO 2002), or either an isolated autosomal monosomy or monosomal karyotype (Breems et al. JCO 2008). This evidence suggests that tumor-specific mechanisms cooperate to overcome the unfitness barrier and maintain aneuploidy. However, the molecular bases of aneuploid AML are incompletely understood.
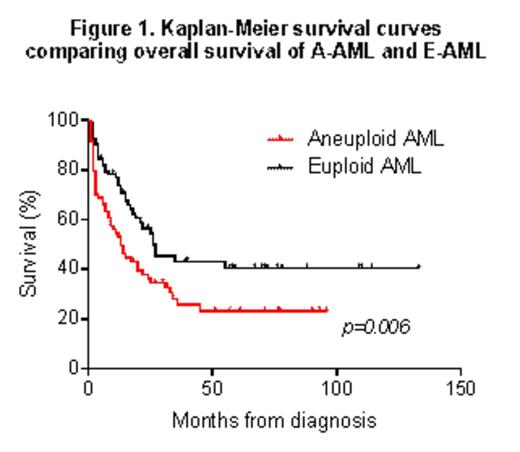
We analyzed a cohort of 166 cytogenetically-characterized AML patients (80 aneuploid (A-) and 86 euploid (E-)) treated at Seràgnoli Institute (Bologna). Aneuploidy was significantly associated with poor overall survival (median survival: 13 and 26 months in A-AML and E-AML respectively; p=.006, Fig.1).
To identify AML-specific alterations having a causative and/or tolerogenic role towards aneuploidy, we integrated high-throughput genomic and transcriptomic analyses.
We performed 100 bp paired-end whole exome sequencing (WES, Illumina Hiseq2000) of 70 samples from our A-AML and E-AML cohort of 166 patients. Variants where called with MuTect or GATK for single nucleotide variant and indels detection, respectively. AML samples were genotyped by CytoScan HD Array (Affymetrix). Gene expression profiling (GEP) was also conducted on bone marrow cells from 24 A-AML, 33 E-AML (≥80% blasts) and 7 healthy controls (HTA 2.0, Affymetrix).
We detected a significantly higher mutation load in A-AML compared with E-AML (median number of variants: 31 and 15, p=.04) which was interestingly unrelated to patients' age (median age: 63.5 years in A-AML and 62 years in E-AML, Xie et al, Nat. Med. 2014). C>A and C>T substitutions, which are likely mediated by endogenous 5mCdeamination, were the most frequent alterations (Alexandrov et al. Nat. 2013). However, aneuploidy associated with an increased variability in terms of mutational signatures, with the majority of A-AML displaying 3 or more signatures compared to few E-AML cases (p=.04).
WES analysis also revealed a specific pattern of somatic mutations in A-AML. A-AML had a lower number of mutations in signaling genes (p=.04), while being enriched for alterations in cell cycle genes (p=.01) compared with E-AML. The mutated genes were involved in different cell cycle phases, including DNA replication (MCM6, PURB, SSRP1), centrosome dynamics (CEP250, SAC3D1, HEPACAM2, CCP110), chromosome segregation (NUSAP1, ESPL1, TRIOBP), mitotic checkpoint (ANAPC7, FAM64A) and regulation (CDK9, MELK, ZBTB17, FOXN3, PPM1D, USP2). Moreover, genomic deletion of cell cycle-related genes was frequently detected in A-AML. Notably, ESPL1 which associated with aneuploidy, chromosome instability and DNA damage in mammary tumors (Mukherjee et al. Oncogene 2014) was mutated and also upregulated in A-AML compared with E-AML (p=.01), the latter showing expression levels comparable to controls. Among the top-ranked genes differentially expressed between A-AML and E-AML, we identified a specific signature characterized by increased CDC20 and UBE2C and reduced RAD50 and ATR in A-AML (p<.001), which has been previously linked to defects in chromosome number. Additional mutations targeting DNA damage and repair pathways were identified in A-AML, including TP53 mutations, which account for 15% of cases. Moreover, A-AML showed a significant upregulation of a KRAS transcriptional signature and downregulation of FANCL- and TP53-related signatures, irrespective of TP53 mutational status.
Our data show a link between aneuploidy and genomic instability in AML. Deregulation of the cell cycle machinery, DNA damage and repair checkpoints either through mutations, copy number and transcriptomic alterations is a hallmark of A-AML. The results define specific genomic and transcriptomic signatures that cooperate with leukemogenic pathways, as KRAS signaling, to the development of the aggressive phenotype of A-AML and suggest that a number of A-AML patients may benefit frompharmacological reactivation of TP53pathway (e.g. MDM2 inhibitor, clinical trial NP28679).
Supported by: FP7 NGS-PTL project, ELN, AIL, AIRC, PRIN, progetto Regione-Università 2010-12
GS & AP: equal contribution
Soverini:Novartis, Briston-Myers Squibb, ARIAD: Consultancy. Cavo:JANSSEN, CELGENE, AMGEN: Consultancy. Haferlach:MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Martinelli:MSD: Consultancy; BMS: Speakers Bureau; Roche: Consultancy; ARIAD: Consultancy; Novartis: Speakers Bureau; Pfizer: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.