Abstract
While survival outcomes after HLA-matched unrelated donor (URD) blood and marrow transplant (BMT) have significantly improved over the last 2 decades, about 40% of patients die of various causes before one-year post-URD BMT. We performed a genome-wide association study (GWAS) named DISCOVeRY-BMT (Determining the Influence of Susceptibility COnveying Variants Related to one-Year mortality after BMT) of overall (OS) and progression-free (PFS) survival in 3,532 patients treated for AML, ALL or MDS and reported to CIBMTR from 2000-2011 and their 8/8 HLA-matched URDs. Cohort 1 consisted of 2,609 patients with 10/10 HLA-matched URD BMT from 2000-08; Cohort 2 consisted of 923 patients with 10/10 HLA-matched URD BMT from 2009-11 and 8/8 (but <10/10) HLA-matched URD BMT from 2000-11. Genotyping on recipients and donors was performed using the HumanOmniExpress-24 BeadChip (Illumina, San Diego, CA), containing approximately 730,000 single nucleotide polymorphisms (SNPs); the CEU reference panel from the 1000 Genomes project (March 2012 release) was used to impute additional genotypes not available on the GWAS chip or that failed typing. Samples were pre-phased with SHAPEIT and genotypes inferred using IMPUTE2. After quality control, Cohort 1 included 2,108 recipients and 2,052 donors and Cohort 2 included 773 recipients and 760 donors of European-American ancestry typed at 637,655 markers and ~9 million imputed SNPs. Each SNP was measured for association with OS and PFS using Cox proportional hazard models in R. All models included age at BMT, diagnosis (AML, ALL, MDS), disease status at BMT (early, intermediate, advanced), cell source (blood, marrow) and year of BMT. P-values were combined using METAL software with weights proportional to the square root of the number of cases. We report results for a combined P -value (Pmeta) <5x10-8.
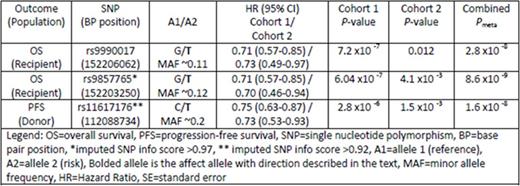
The T allele of rs9990017 in recipients is significantly associated with better OS (Table) yielding about a 30% decreased hazard of all causes of death including disease relapse, graft-versus-host disease, infection and organ failure. Analysis of the effect of donor genotype at this SNP shows no evidence of association with OS or PFS. The strongest imputed association in this region (info score >.98) was rs9857765 with the T allele at this locus conferring a significantly improved OS in both cohorts (Table).
Two SNPs (rs9845520 and rs9839074) in strong linkage disequilibrium (r2 =.94) with both rs9857765 and rs9990017, are predicted to affect transcription factor binding (Regulomedb score=2b) of the highly-conserved gene MBNL1 and the common allele in rs9839074 is predicted to be in an important position in the GATA1 (erythroid development) motif and a binding site for CEBPB, STAT3 and FOS across multiple cell lines. MBNL1 protein is expressed in most blood and marrow hematopoietic cells, while MBNL1 mRNA is expressed in many organs. CEBPB regulates immune response genes, is required for normal macrophage function/differentiation and can interact with (among others) glucocorticoid receptor, IL-6, TNF-alpha and transporter proteins conferring multi-drug resistance (ABCC2, ABCB1). Normal activation of STAT3 is required for self-renewal of embryonic stem cells and differentiation of TH17 cells, while constitutive activation of STAT3 is associated with poor prognosis in acute leukemia. FOS is involved with CD16+ signaling in NK cells, vitamin D receptor gene regulation in osteoporosis, and proliferation of hematopoietic cells, among other roles.
A region on 13q34 showed evidence of association with PFS; recipients whose donor had a T allele at rs11617176 showed improved PFS compared to those whose donors had the C allele (see Table). This region contains variants identified in other GWAS of mean platelet volume, coronary artery disease, interstitial lung disease, response to tocilizumab in rheumatoid arthritis patients and renal transplant outcomes, among others.
Our study, DISCOVeRY-BMT, is the first GWAS for URD BMT survival outcomes. Confirmation of these findings in a third cohort, genotyping of imputed SNPs, and further studies of the functional consequences of these SNPs may provide more individualized risk prediction and prognosis, while confirmation of rs11617176 may aid in donor selection.
Hahn:Novartis: Equity Ownership; NIH/NHLBI: Research Funding. McCarthy:Celgene: Honoraria, Membership on an entity's Board of Directors or advisory committees; Sanofi: Honoraria, Membership on an entity's Board of Directors or advisory committees; Bristol Myers Squibb: Honoraria, Membership on an entity's Board of Directors or advisory committees; The Binding Site: Honoraria, Membership on an entity's Board of Directors or advisory committees; Karyopharm: Honoraria, Membership on an entity's Board of Directors or advisory committees; Onyx: Honoraria, Membership on an entity's Board of Directors or advisory committees; Janssen: Honoraria, Membership on an entity's Board of Directors or advisory committees. Sucheston-Campbell:NIH/NHLBI: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.