Abstract
BACKGROUND. Recent studies using whole genome approaches have shown that acquired mutations in genes such as DNMT3A, TET2, ASXL1, JAK2 and TP53 are observed in a subset of elderly subjects without hematologic malignancies, and are associated with an increased relative risk of developing myeloid cancers, including acute myeloid leukemia (AML) and myelodysplastic syndromes (MDS). The goal of this study is to precisely determine, using a more sensitive approach, the prevalence of somatic mutations and their allelic frequencies in elderly subjects, and assess the impact of somatic mutations on biological characteristics.
METHODS. Cohort: 1101 hematologically healthy women aged 70 to 101 years old were recruited from the general population. All patients in the cohort were characterized with a i) medical questionnaire; ii) complete blood counts (CBC), and iii) X-chromosome inactivation (XCI) patterns in polymorphonuclear (PMN), using HUMARA to assess the clonality of myeloid-derived cells. Sequencing: All individuals were sequenced using a targeted approach at high coverage (95% >500x) on an Ion Proton NGS instrument. Libraries were generated using an amplicon-based panel covering 19 recurrently mutated genes in myeloid malignancies (ASXL1, BRAF, CBL, CEBPA, DNMT3A, FLT3, GATA2, IDH1, IDH2, JAK2, KIT, KRAS, NPM1, NRAS, PTPN11, RUNX1, TET2, TP53 and WT1) with 237 amplicons over 22kb. Statistical analyses: Mutational status was correlated with the biological characteristics of subjects using linear regression analysis (GLM). All age-dependent variables were corrected for age (XCI skewing PMN↑, monocytes↑, hemoglobin↓, lymphocytes↓ and platelets↓).
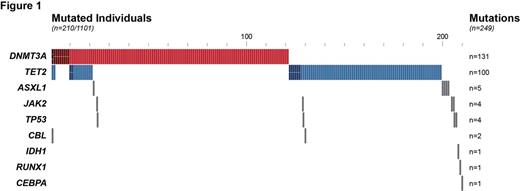
RESULTS. Mutations. We identified 249 somatic mutations in 210 of the 1101 individuals (19%) with variant allele frequencies (VAF) ranging from 3.6% to 74.8%. Mutations were found in DNMT3A (n=131), TET2 (n=100), ASXL1 (n=5), JAK2 (N=4), TP53 (n=4), CBL (n=2), IDH1 (n=1), RUNX1 (n=1) and CEBPA (n=1). TET2 and DNMT3A mutations accounted for 93% of all somatic mutations. More than one somatic mutation was observed in 32 individuals (3%), with one subject harboring 5 mutations. Correlation between mutational status and biological characteristics. The presence of mutations significantly increased with age (all genes: P =0.00024, TET2: P =0.00016, TET2 and DNMT3A (double mutations): P =0.03268), although no age effect was documented for DNMT3A mutation alone. The presence of a mutation in TET2, but not in other disease alleles correlated with increased myeloid skewing (P= 0.0008). The presence of a mutation (all genes) or a mutation in DNMT3A correlated with a decreased mean corpuscular volume (MCV) (P= 0.046). TET2 mutations were associated with decreased total white blood cell counts (P =0.0262) and PMN counts (P =0.0137). DNMT3A correlated with increased total white blood cell counts (P =0.035) and increased lymphocyte counts (P =0.0256). No mutational phenotype correlated with alterations in monocyte counts. Despite these global mutation-associated hematological phenotypes, we observed several individuals with a high VAF (>30%) in DNMT3A or TET2 with completely normal CBC.
CONCLUSION: Acquired mutations in the normal aging population occur at very high frequency, most commonly TET2 and DNMT3A. This suggests that mutation-driven epigenetic dysregulation is key in the development of age-associated hematological cancers. TET2 and DNMT3A mutations are associated with distinctive characteristics such as increased clonal dominance, aging and lower PMN counts for TET2; increased lymphocyte counts and decreased MCV for DNMT3A. This suggests that these two genes may have a different impact on transformation to malignant phenotype. Prospective evaluation of this cohort and sequential analysis of blood specimens will provide informative insight on the sequence of events leading to age-associated hematological cancers.
Mollica:Pfizer: Consultancy; BMS: Consultancy; Novartis: Consultancy. Busque:Novartis: Consultancy; BMS: Consultancy; PFIZER: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.