Abstract
Background: Successful outcome in childhood acute lymphoblastic leukemia (ALL) relies upon appropriate central nervous system (CNS)-directed therapy for treatment of subclinical or overt CNS leukemia. Patients with leukemia blasts detected in the cerebrospinal fluid (CSF) at diagnosis have poorer survival compared with patients without blasts in the CSF and require intensified intrathecal therapy to avert a higher rate of relapse. Traditionally, CSF assessment is performed by morphological analysis of CSF smears and the blasts confirmed by immunohistochemistry or flow cytometry. The confirmation tests are done at few institutions and rely on subjective operator judgment for identification of leukemic blasts. A precise definition of CNS involvement is necessary to avoid over- or under- treatment. Next-generation sequencing (NGS) is currently being used for minimal residual disease (MRD) assessment in lymphoid malignancies (Faham et al., Blood 2012). In this study, we assessed whether the NGS method could be used to detect leukemic clonotypes in CSF samples from 89 newly diagnosed pediatric ALL patients.
Methods: Diagnostic bone marrow samples and paired CSF samples were obtained from 89 patients. CSF samples were collected in one tube and fractionated for Wright stained cytospins (1ml), TdT immunohistochemistry (1ml), and the remainder (3-5ml) for NGS-based MRD studies (Adaptive Biotechnologies, South San Francisco, CA). Briefly, using universal primer sets, we amplified variable, diversity, and joining gene segments from immunoglobulin (Ig) heavy chain (IGH), Ig kappa chain (IGK), and T-cell receptor beta (TRB), delta (TRD) and gamma (TRG) loci from genomic DNA. Amplified products were sequenced and analyzed using standardized algorithms for clonotype determination. Tumor-specific clonotypes were identified for each patient based on their high-frequency within the B-cell repertoire. The presence of the tumor-specific clonotype was then quantitated in CSF (cell pellet and supernatant) samples obtained at diagnosis. A quantitative and standardized measure of leukemic clonotype level per million leukocytes in each follow-up sample was determined.
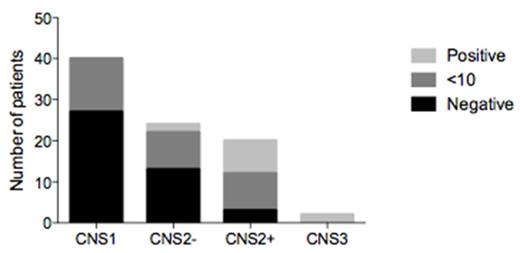
Results: Diagnostic bone marrow samples from 89 pediatric ALL patients were used to identify the tumor-specific clonotypes. At least one tumor-specific clonotype was identified in 86/89 (97%) of the patients. The IGH-VDJ assay was the most frequent gene rearrangement: at least one IGH-VDJ clonal rearrangement was detected in 66 of the diagnostic ALL samples. TRD was the second most informative receptor with a clonal rearrangement found in 43 patients, followed by TRG in 38 patients, IGK in 31 patients, TCB in 16 patients and IGH-DJ in 13 patients. The level of tumor burden associated with each leukemic clonotype was then assessed in CSF samples drawn at diagnosis from each of the 86 patients. All 40 CNS1 patients had no detectable or minimal (<10) leukemic cell equivalents. Of the 24 patients with CNS2 but TDT negative status, only 2 had >10 leukemic cell equivalents in the CSF. By contrast, 8 of 20 (40%) patients with CNS2 and TDT positive status and both patients with CNS3 had CSF with >10 leukemic cell equivalents (Figure 1).
Conclusions: This study demonstrates that the NGS-based method can be used to assess CNS involvement in pediatric patients with ALL. Based on these results, NGS-based CNS assessment will be used for patient stratification in future clinical trials for pediatric ALL.
Zheng:Adaptive Biotechnologies Corp.: Employment, Equity Ownership. Carlton:Adaptive Biotechnologies Corp.: Employment, Equity Ownership. Faham:Adaptive Biotechnologies Corp.: Employment, Other: Stockholder.
Author notes
Asterisk with author names denotes non-ASH members.