Abstract

The proteasome inhibitor Bortezomib (Bz), the first agent of a new class of drugs in Multiple Myeloma (MM), has shown remarkable activity and forms an integral part of modern MM treatment. Nevertheless, resistance to Bz eventually develops in a significant proportion of patients, with adverse effects on survival. Numerous publications have addressed this issue through in vitro developed models of acquired Bz resistance (BzR). However the results were quite different in each publication, none of the produced Bz myeloma cell lines was provably stable, no common mechanism of resistance could be demonstrated, and hence were of minimal relevance to the clinical setting.
In order to address these issues an effort was made for the development of an in vitro model of acquired BzR that would resemble the clinical reality in the most accurate way. Two myeloma cell lines were used, one resembling a multisensitive (JJN3) and the other a multiresistant (U266) drug behavior, that were both sensitive to Bz. An at least 20 fold increase in the 48h Bz IC50 was noted for both cell lines. The increase in the IC50 was able to be verified a year after culturing the cell lines in normal medium thus ensuring a stable resistance phenotype.
To delineate the molecular mechanisms that underlie the development of BzR a combined genetic/Gene Expression Profile (GEP) and functional/Proteomics approach was used with emphasis in the common elements of both cell lines. The hypothesis was that if certain pathways are activated in the cells that actually produce the phenotype of BzR they must fulfil two important criteria: 1) They must be present in all the levels of the BzR, 2) The gene changes have to be verified in the level of the gene encoded proteins thus securing their functional importance.
GEP of the naïve cell lines along with the GEP of the Bz resistant cells at different levels of BzR (5-fold, 10-fold, 20-fold) were used. The statistical analysis revealed 100 gene probes common in both cell lines that achieved their highest change as soon as BzR was established and remained stable at that level for all later versions (P<0.1, q<0.1) and 115 gene probes common in both cell lines that their change was proportional to the level of BzR (P<0.001, q <0.005).
The proteomics analysis of the Bz resistant cell lines at their latest level of resistance (20-fold) revealed 262 proteins common in both cell lines that were up-regulated and 263 common in both cell lines that were down-regulated (change >10% to be considered significant).
The intersection of the list of the common genes with the list of the common proteins revealed 47 gene-proteins all but one novel in MM. They can be grouped in distinct biological categories with the most prominent ones being the ROS/Mitochondrial Factor category comprising of 10 gene-proteins, the E3 Ubiquitin Pathway 6 genes-proteins and Translation Regulation 5 genes-proteins.
Even more importantly 30 of them have profound survival implications in MM -all of them novel in MM- both for Overall Survival (OS) and Progression Free Survival (PFS) in both Bz (TT3) and non Bz (TT2) containing protocols implying that myeloma cells apply both Bz specific and non-specific mechanisms to acquire BzR.
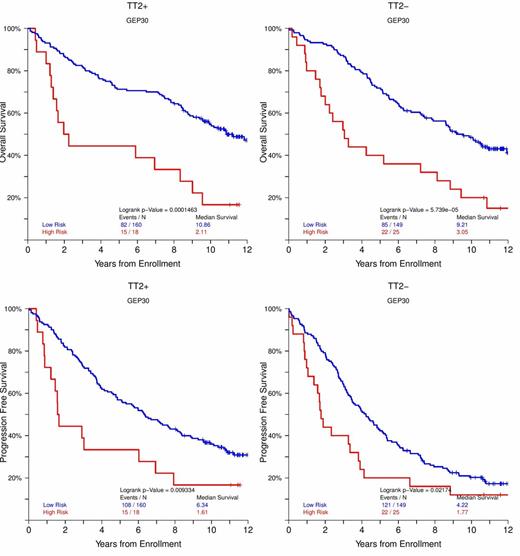
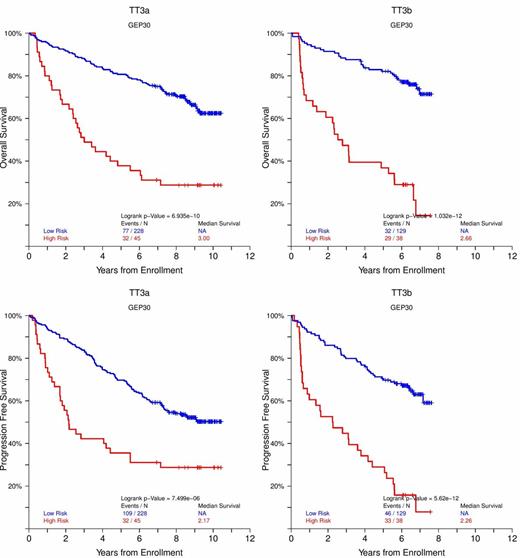
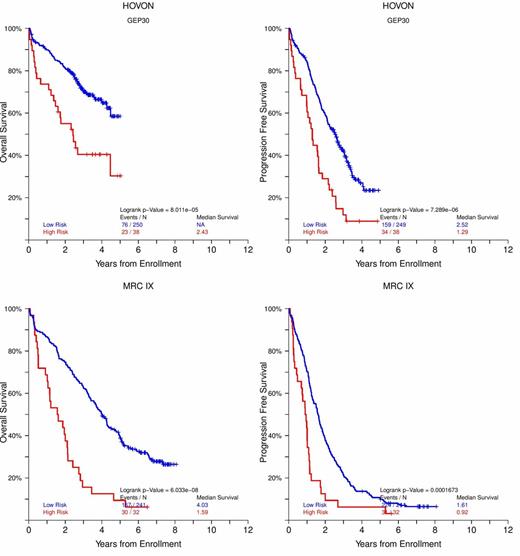
Based on these 30 genes-proteins a GEP risk score (GEP-30) was constructed that was able to achieve remarkable statistical power in both Bz containing and non containing trials of both newly diagnosed (TT2 with and without thalidomide i.e. TT2+ and TT2-, TT3a, TT3b, HOVON, MRC IX, Figure 1A,B,C) and relapsed MM (TT6 , OS: NR vs 1.52 yr P<0.00001, PFS: NR vs 1.13 yr P<0.00001 for low and high risk)
KM plots for OS and PFS of GEP-30 for newly diagnosed MM
KM plots for OS and PFS of GEP-30 for newly diagnosed MM
Stein:University of Arkansas for Medical Sciences: Employment. Barlogie:University of Arkansas for Medical Sciences: Employment. Epstein:University of Arkansas for Medical Sciences: Employment. Heuck:Janssen: Other: Advisory Board; Celgene: Consultancy; Millenium: Other: Advisory Board; Foundation Medicine: Honoraria; University of Arkansas for Medical Sciences: Employment.
Author notes
Asterisk with author names denotes non-ASH members.

This icon denotes a clinically relevant abstract