Key Points
Tmod3-null embryos have macrothrombocytopenia due to impaired MK cytoplasmic morphogenesis with defective proplatelet formation.
F-actin polymerization and organization are disrupted in Tmod3-null MKs and in their proplatelet buds.
Abstract
The actin cytoskeleton is important for platelet biogenesis. Tropomodulin-3 (Tmod3), the only Tmod isoform detected in platelets and megakaryocytes (MKs), caps actin filament (F-actin) pointed ends and binds tropomyosins (TMs), regulating actin polymerization and stability. To determine the function of Tmod3 in platelet biogenesis, we studied Tmod3−/− embryos, which are embryonic lethal by E18.5. Tmod3−/− embryos often show hemorrhaging at E14.5 with fewer and larger platelets, indicating impaired platelet biogenesis. MK numbers are moderately increased in Tmod3−/− fetal livers, with only a slight increase in the 8N population, suggesting that MK differentiation is not significantly affected. However, Tmod3−/− MKs fail to develop a normal demarcation membrane system (DMS), and cytoplasmic organelle distribution is abnormal. Moreover, cultured Tmod3−/− MKs exhibit impaired proplatelet formation with a wide range of proplatelet bud sizes, including abnormally large proplatelet buds containing incorrect numbers of von Willebrand factor-positive granules. Tmod3−/− MKs exhibit F-actin disturbances, and Tmod3−/− MKs spreading on collagen fail to polymerize F-actin into actomyosin contractile bundles. Tmod3 associates with TM4 and the F-actin cytoskeleton in wild-type MKs, and confocal microscopy reveals that Tmod3, TM4, and F-actin partially colocalize near the membrane of proplatelet buds. In contrast, the abnormally large proplatelets from Tmod3−/− MKs show increased F-actin and redistribution of F-actin and TM4 from the cortex to the cytoplasm, but normal microtubule coil organization. We conclude that F-actin capping by Tmod3 regulates F-actin organization in mouse fetal liver-derived MKs, thereby controlling MK cytoplasmic morphogenesis, including DMS formation and organelle distribution, as well as proplatelet formation and sizing.
Introduction
Blood platelets arise from megakaryocyte (MK) precursors.1 During maturation, MKs become polyploid and grow up to 50 to 100 μm in size, forming a well-organized demarcation membrane system (DMS) (also termed invaginated membrane system) in their cytoplasm, defining the cytoplasmic domains of platelet territories, and providing the membrane reservoir for release into proplatelet protrusions.2,3 Released proplatelets are then processed into mature platelets in the circulation.4,5 Platelet biogenesis relies on cytoskeletal rearrangements of both microtubules and actin filament (F-actin) networks.2,3,6,7 Microtubule assembly and dynein-mediated microtubule sliding are important for proplatelet elongation and organelle trafficking into proplatelet extensions,2,6,8,9 whereas F-actin plays a role in numbers and branching of proplatelet protrusions,1 and their morphology, size, and release from the proplatelet extensions.2,3,7 Analyses of human mutations and gene-targeted mice have identified several actin cytoskeletal components involved in this process,7 including nonmuscle myosin IIA (NMIIA) heavy chain (MYH9),10-13 actin depolymerizing factor (ADF)/n-cofilin,14 filamin A (FLNA),15,16 Wiskott-Aldrich syndrome protein (WASp),17 Rho GTPases,18,19 the formin DIAPH1 (mDia1),20 and α-actinin1.21,22 Human mutations and mouse knockouts for these actin cytoskeletal factors often result in thrombocytopenias with abnormally sized and poorly functioning platelets, as well as aberrant MKs characterized by perturbations in F-actin organization and impaired formation of proplatelet extensions. However, MK numbers and polyploidy are relatively unaffected by mutations in these cytoskeletal proteins.11,15,18
In addition to defects in F-actin organization, a common feature underlying the MK phenotypes of some of these mutants is an inability to elaborate the DMS that defines the cytoplasmic territories destined to become platelets and supplies the extensive membrane reservoir for proplatelet formation.23 This is particularly evident in Myh9−/− (NMIIA-null) mouse MKs, which have an uneven organelle distribution and fragmentary DMS with defective F-actin organization.10,11 Patel-Hett et al have demonstrated that the DMS is supported by a spectrin-actin cytoskeleton,24 similar to platelet membranes.25 Transduction of mouse MKs with a spectrin tetramer-disrupting construct inhibits formation of the DMS, impairs formation of proplatelet extensions, and leads to loss of the characteristic barbell shape of newly formed proplatelets.24 Actomyosin contraction and spectrin-actin networks both rely upon F-actin stability, yet other studies have demonstrated that platelet biogenesis depends upon F-actin disassembly and turnover promoted by ADF/n-cofilin.14 To reconcile these apparently contradictory roles for F-actin dynamics, we hypothesize that a balance between F-actin stability and turnover may be critical for platelet biogenesis, with F-actin stabilizing proteins in MKs counteracting F-actin disassembly-promoting proteins such as ADF/n-cofilin.
Tropomodulins (Tmods) play key roles in stabilizing F-actin networks by binding tropomyosins (TMs) and capping the pointed ends of TM-associated F-actin.26 TMs bind along the length of F-actin and inhibit disassembly by ADF/cofilin or severing by gelsolin, and TM-mediated F-actin stabilization is enhanced by Tmods.27-33 Tmods are components of membrane-associated TM–F-actin networks, including the spectrin-actin network of the erythrocyte membrane (Tmod1), ocular lens fiber cell membranes (Tmod1), polarized epithelial cell plasma membranes (Tmod3), and the sarcoplasmic reticulum membranes of skeletal muscle (Tmod3), where Tmod-mediated stabilization of TM–F-actin is critical for membrane morphology, mechanics, and physiology.34-37 Tmod3 is also involved in endothelial cell migration,38 erythroblast enucleation and erythroblast-macrophage adhesion in erythropoiesis,39 and insulin-stimulated GLUT4 trafficking in adipocytes.40
In this study, we show that Tmod3 is the only Tmod in platelets and MKs and confirm that TM4 (Tpm4.2, encoded by the Tpm4/δTm gene41 ) is the predominant TM in MKs and platelets.23,42 We investigated a role for Tmod3 in platelet formation by studying Tmod3−/− mice, which are embryonic lethal at E14.5 to E18.5 with anemia due to impaired fetal liver erythropoiesis.39 Tmod3−/− embryos also display hemorrhages, which may be partly due to reduced platelet numbers and enlarged platelets in the embryonic circulation. Tmod3−/− fetal liver MKs have reduced proplatelet formation in vitro, but no significant changes in MK ploidy and a moderate increase in MK numbers. However, Tmod3−/− MK cytoplasmic morphogenesis is impaired, with insufficient DMS formation and aberrant organelle distribution, and many proplatelet buds from cultured MKs are abnormally large with incorrect numbers of von Willebrand factor (VWF) negative and positive granules. Tmod3 associates with TM4 and the actin cytoskeleton in MKs, and absence of Tmod3 leads to defective F-actin polymerization and organization in MKs and proplatelet buds. Deletion of Tmod3 may disrupt F-actin in MKs via perturbation of TM4–F-actin interactions, resulting in F-actin instability and reorganization, leading to defective DMS formation and organelle distribution, thereby impairing proplatelet formation, morphology, and organelle content.
Materials and methods
Mice
Tmod3−/− embryos were as described.39 All experiments were performed according to the National Institutes of Health animal care guidelines, as approved by The Scripps Research Institute’s Institutional Animal Care and Use Committee.
X-gal and hematoxylin and eosin (H&E) staining
E14.5 embryos were subjected to whole-mount X-gal staining as described.39 Embryos were then fixed, paraffin-embedded, and sectioned for H&E staining.
In vitro culture of MKs
Dissociated cells from E13.5 to E14.5 mouse fetal livers were cultured in Dulbecco’s Modified Eagle’s medium (Gibco Life Technologies) with 5 ng/mL recombinant murine thrombopoietin (mTPO, Life Technologies), 10% fetal bovine serum (HyClone, Thermo Scientific), penicillin, and streptomycin.43 MKs were collected using a 2-step bovine serum albumin (BSA) density gradient on day 3 and incubated in Dulbecco’s Modified Eagle’s medium with mTPO for an additional 24 hours until proplatelet formation.43
Immunofluorescence and confocal microscopy
Whole mouse embryos were processed for cryosectioning and labeled for antibody as described.39 For imaging of fetal liver cytospins, E14.5 fetal livers were dissociated into Dulbecco’s phosphate-buffered saline (PBS), suspended cells were fixed in 4% paraformaldehyde overnight at 4°C, permeabilized, blocked, and labeled with antibodies in suspension, and then spun onto slides.39 For staining of proplatelet-forming MKs from in vitro cultures, cells were fixed in 4% paraformaldehyde for at least 30 minutes at room temperature and then centrifuged onto poly-l-lysine coated slides.43 Cells or tissue preparations were permeabilized in 0.3% Triton X-100 in PBS and blocked overnight at 4°C in 2% BSA with 1% goat serum in PBS + 0.1% Triton X-100. Slides were incubated with primary antibodies (see supplemental Table 1 on the Blood Web site) overnight at 4°C, followed by secondary antibodies (supplemental Table 1) for 2 hours at room temperature, and mounted in ProLong Gold antifade reagent (Life Technologies). Images were acquired using a Zeiss LSM 780 confocal laser scanning fluorescence microscope, and analyzed using Volocity 6.3 (PerkinElmer) or ImageJ software as indicated in figure legends.
Transmission electron microscopy (TEM)
Peripheral blood from E14.5 mouse embryos was collected by allowing embryos to bleed out into Dulbecco’s PBS with 5 mM EDTA and added to K3EDTA containing tubes (Becton Dickinson). Blood or dissociated fetal liver cells were fixed overnight in ice-cold 4% paraformaldehyde and 1.5% glutaraldehyde in 0.1 M sodium cacodylate at pH 7.2. Cells or tissues were processed for embedding and thin sectioning as described.44 Sections were examined on a Philips CM100 electron microscope (FEI). Images were collected using a MegaView III CCD camera (Olympus Soft Imaging Solutions GmbH).
Ploidy analysis
Dissociated E14.5 fetal liver cells were cultured in StemSpan Serum-Free Expansion Medium (Stemcell Technologies) supplemented with 30% BSA, insulin, and transferrin (Stemcell Technologies) and 50 ng/mL mTPO (ConnStem) for 4 days at 37°C. Cultured cells were harvested for flow cytometry as described (supplemental Figure 2).45
Western blotting
Mouse blood was collected via cardiac puncture of 3-month-old male mice directly into K3EDTA Vacutainer tubes (Becton Dickinson). Human peripheral blood from healthy donors was collected by venipuncture into K3EDTA according to an Institutional Review Board–approved human subjects blood collection protocol (11-5773). Red blood cells and leukocytes were removed by centrifugation at 200g for 10 minutes, followed by collection of platelets from the plasma by centrifugation at 1000g for 5 minutes. MKs were obtained from in vitro cultured mouse fetal liver cells on day 3 as described above. Western blotting was performed as described.39 Primary and secondary antibodies are shown in supplemental Table 1.
Co-immunoprecipitation (co-IP)
Cultured MKs were lysed at 4°C in modified radioimmunoprecipitation assay buffer (50 mM Tris pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.1% sodium dodecyl sulfate, and 1% NP40) supplemented with protease inhibitor cocktail (1:100, Sigma-Aldrich), centrifuged for 10 minutes at 4°C in an Eppendorf microcentrifuge to remove insoluble material, and affinity-purified rabbit anti-Tmod3 (1 µg), rabbit anti-TM4, or pre-immune control rabbit IgG (1 µg) was added. After incubation for 1 hour, μMACS Protein A MicroBeads (Miltenyi Biotec) was added, incubated overnight at 4°C, run through MACS separation columns (Miltenyi Biotec), and antibody-protein complexes were eluted with sodium dodecyl sulfate-gel sample buffer. Western blotting of co-IP products and input whole-cell lysate were then performed, with images acquired with an Odyssey Imager (LI-COR Biosciences).46
Statistical analysis
Data are presented as mean ± SEM for small sample sizes (<10) and mean ± SD for large sample sizes (>30). A 2-tailed unpaired Student t test was used for data analysis. P < .05 was considered statistically significant. Prism Version 5 software (GraphPad) and Microsoft Excel were used for statistical analysis.
Results
Tmod3 is present in platelets and MKs
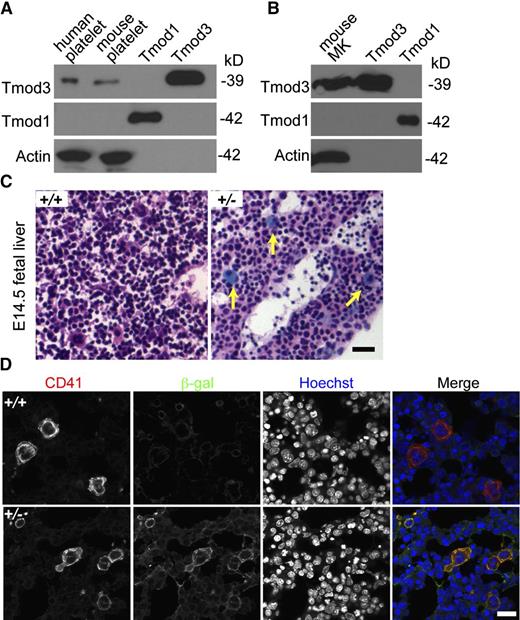
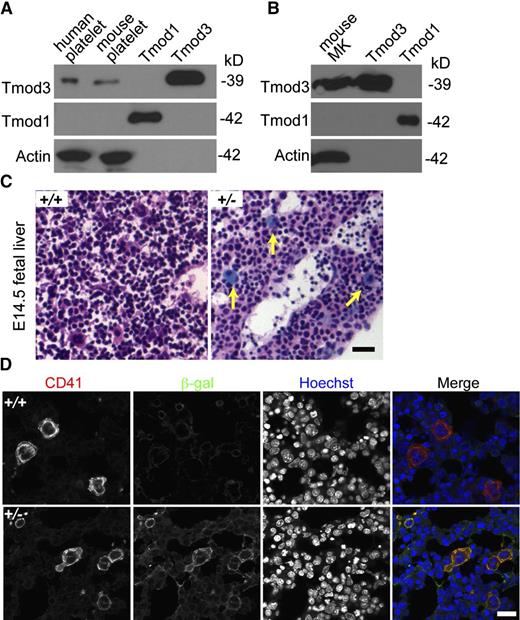
Tmod3 is the only Tmod isoform in the human platelet proteome, with an estimated 11 900 copies per platelet, considerably less abundant than cofilin (CFL1 + CFL2) at 337 200 copies, or profilin (PFN1) at 504 000 copies per platelet.42 Western blotting confirms that Tmod3 is present in both human and mouse platelets, whereas no Tmod1 is detected (Figure 1A). Similar results are obtained using in vitro cultured MKs from E13.5 mouse fetal livers (Figure 1B). LacZ staining of fetal livers from Tmod3+/− mice39 also reveals Tmod3 expression in large multinucleated MKs (Figure 1C), along with fainter blue staining in other cells, as previously shown.39 Immunostaining for CD41 and β-gal confirmed abundant β-gal in CD41+, multinucleated MKs (Figure 1D). Indeed, Tmod3 expression appears to be higher in MKs in comparison with other fetal liver cell types. Based on the restricted expression of Tmod2 and Tmod4 in neurons and skeletal muscle, respectively,26 Tmod3 is the only Tmod in platelets and fetal liver MKs.
Tmod3 is expressed in platelets and MKs. (A) Western blotting of Tmod expression in human and mouse platelets. (B) Western blotting of Tmods in MKs cultured from mouse fetal liver (E13.5); 10 ng purified recombinant Tmod1 and Tmod3 proteins were used as controls (A-B). (C) LacZ staining of sections from Tmod3+/+ (left) and Tmod3+/− (right) E14.5 embryos. Embryos were dissected and stained with X-gal and H&E. Yellow arrows indicate large MKs with bright blue staining. Bar, 40 µm. Images were acquired with a ×20 objective (N.A. 0.5) using a Zeiss Axioskop microscope and a Zeiss AxioCam ICc3 color camera. (D) Immunofluorescence staining for β-galactosidase (green) and CD41 (red) in cryosections of Tmod3+/+ and Tmod3+/− E14.5 embryos. Bar, 20 µm. Images were acquired using a Zeiss LSM 780 confocal laser scanning fluorescence microscope with a ×60 oil-immersion objective (N.A. 1.4).
Tmod3 is expressed in platelets and MKs. (A) Western blotting of Tmod expression in human and mouse platelets. (B) Western blotting of Tmods in MKs cultured from mouse fetal liver (E13.5); 10 ng purified recombinant Tmod1 and Tmod3 proteins were used as controls (A-B). (C) LacZ staining of sections from Tmod3+/+ (left) and Tmod3+/− (right) E14.5 embryos. Embryos were dissected and stained with X-gal and H&E. Yellow arrows indicate large MKs with bright blue staining. Bar, 40 µm. Images were acquired with a ×20 objective (N.A. 0.5) using a Zeiss Axioskop microscope and a Zeiss AxioCam ICc3 color camera. (D) Immunofluorescence staining for β-galactosidase (green) and CD41 (red) in cryosections of Tmod3+/+ and Tmod3+/− E14.5 embryos. Bar, 20 µm. Images were acquired using a Zeiss LSM 780 confocal laser scanning fluorescence microscope with a ×60 oil-immersion objective (N.A. 1.4).
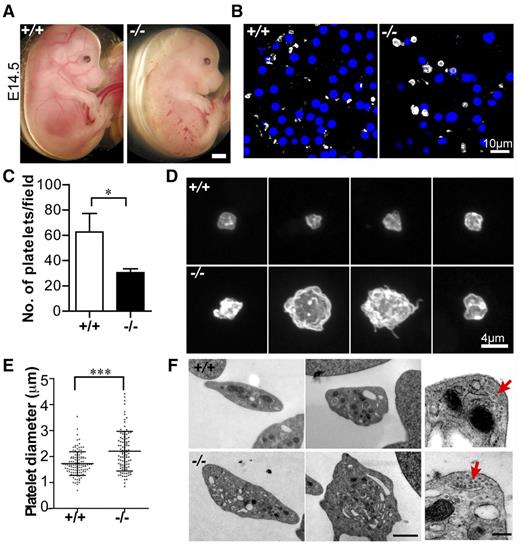
Tmod3−/− embryos have hemorrhages and fewer platelets with abnormal morphology
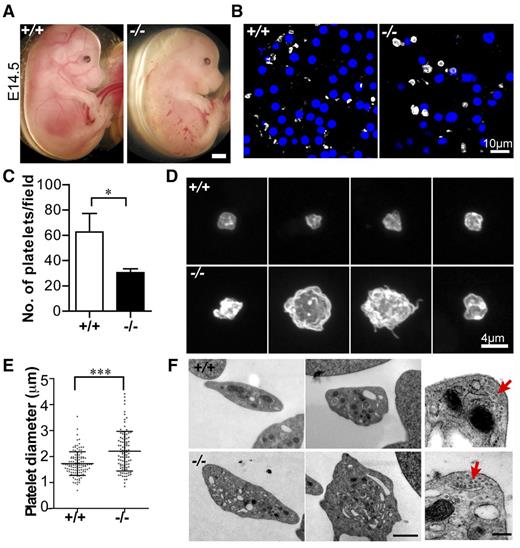
We previously noticed small hemorrhages in ∼75% of Tmod3−/− embryos at E14.5 (Figure 2A).39 As Tmod3 is expressed in endothelial cells38,39 and platelets (Figure 1), hemorrhages may be due to endothelial barrier and/or platelet dysfunction. We focused on the platelet phenotype, because Tmod3−/− embryos had fewer circulating platelets (∼50% of wild-type [WT]) (Figure 2B,C), with larger and more variable sizes (Figure 2B-E). Tmod3−/− platelets were also more irregular in shape and often displayed small protrusions, unlike WT platelets (Figure 2D). TEM revealed more vesicular structures with clear lumens in some Tmod3−/− platelets (Figure 2F), and confocal microscopy of GPIbα revealed internal staining in contrast to typical membrane staining of WT platelets (supplemental Figure 1). This could be due to partial activation of Tmod3−/− platelets leading to GPIbα internalization47 or impaired membrane biogenesis (see below in Figures 4 and 5). However, no changes were evident in the morphology of the circumferential microtubule coils of Tmod3−/− platelets (Figure 2F, red arrows). (Tmod3+/− embryos and platelets were similar to WT, due to increased Tmod3 expression in heterozygotes.39 ) Therefore, Tmod3−/− embryos exhibit decreased numbers of platelets with variable sizes and abnormal morphologies, resembling a macrothrombocytopenia.
Tmod3−/− embryos show hemorrhages with abnormal platelets. (A) Representative images of whole Tmod3+/+ (left) and Tmod3−/− (right) embryos at E14.5. Bar, 1 mm. (B) Immunofluorescence staining for CD41 (gray) and Hoechst (blue) in cryosections of blood vessels of Tmod3+/+ (left) and Tmod3−/− (right) embryos, revealing circulating CD41+ platelets in situ. Bar, 10 µm. Images are single optical sections acquired using a Zeiss LSM 780 confocal laser scanning fluorescence microscope with a ×100 oil-immersion objective (N.A. 1.4), zoom 1. (C) Relative numbers of platelets in peripheral blood of Tmod3+/+ and Tmod3−/− E14.5 embryos. Tmod3+/+, 63 ± 20 (n = 4 fields from 2 Tmod3+/+ embryos); Tmod3−/−, 30 ± 7 (n = 5 fields from 2 Tmod3−/− embryos). *P < .05. (D) Representative images of Tmod3+/+ (top) and Tmod3−/− (bottom) platelets stained with CD41. Bar, 4 µm. Images are compressed Z stacks of optical sections acquired using a Zeiss LSM 780 confocal laser scanning fluorescence microscope with a ×100 oil-immersion objective (N.A. 1.4), zoom 3. (E) Platelet sizes in peripheral blood of Tmod3+/+ and Tmod3−/− embryos at E14.5, measured from images as in (B). Platelet diameters were determined from line scans across platelets using Volocity 6.3 software. Tmod3−/− average platelet diameters were ∼×1.5 greater than Tmod3+/+ platelets. Tmod3+/+, 1.73 ± 0.46 µm (n = 114); Tmod3−/−, 2.21 ± 0.76 µm (n = 98). ***P < .001. (F) Representative TEM images of Tmod3+/+ and Tmod3−/− platelets (left and middle panels; Bar, 1 µm), with high magnification views of circumferential microtubule rings (red arrows, right panel; Bar, 200 nm). Average WT platelet diameter in TEM images is ∼3 µm, similar to previous studies,10 but larger than measurements from fluorescence optical sections (B), which include measurements across the short and long axes of the asymmetric platelets (E).
Tmod3−/− embryos show hemorrhages with abnormal platelets. (A) Representative images of whole Tmod3+/+ (left) and Tmod3−/− (right) embryos at E14.5. Bar, 1 mm. (B) Immunofluorescence staining for CD41 (gray) and Hoechst (blue) in cryosections of blood vessels of Tmod3+/+ (left) and Tmod3−/− (right) embryos, revealing circulating CD41+ platelets in situ. Bar, 10 µm. Images are single optical sections acquired using a Zeiss LSM 780 confocal laser scanning fluorescence microscope with a ×100 oil-immersion objective (N.A. 1.4), zoom 1. (C) Relative numbers of platelets in peripheral blood of Tmod3+/+ and Tmod3−/− E14.5 embryos. Tmod3+/+, 63 ± 20 (n = 4 fields from 2 Tmod3+/+ embryos); Tmod3−/−, 30 ± 7 (n = 5 fields from 2 Tmod3−/− embryos). *P < .05. (D) Representative images of Tmod3+/+ (top) and Tmod3−/− (bottom) platelets stained with CD41. Bar, 4 µm. Images are compressed Z stacks of optical sections acquired using a Zeiss LSM 780 confocal laser scanning fluorescence microscope with a ×100 oil-immersion objective (N.A. 1.4), zoom 3. (E) Platelet sizes in peripheral blood of Tmod3+/+ and Tmod3−/− embryos at E14.5, measured from images as in (B). Platelet diameters were determined from line scans across platelets using Volocity 6.3 software. Tmod3−/− average platelet diameters were ∼×1.5 greater than Tmod3+/+ platelets. Tmod3+/+, 1.73 ± 0.46 µm (n = 114); Tmod3−/−, 2.21 ± 0.76 µm (n = 98). ***P < .001. (F) Representative TEM images of Tmod3+/+ and Tmod3−/− platelets (left and middle panels; Bar, 1 µm), with high magnification views of circumferential microtubule rings (red arrows, right panel; Bar, 200 nm). Average WT platelet diameter in TEM images is ∼3 µm, similar to previous studies,10 but larger than measurements from fluorescence optical sections (B), which include measurements across the short and long axes of the asymmetric platelets (E).
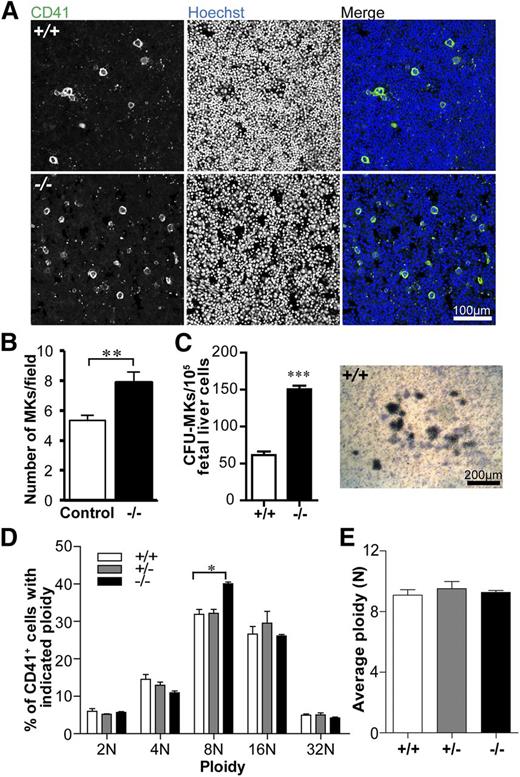
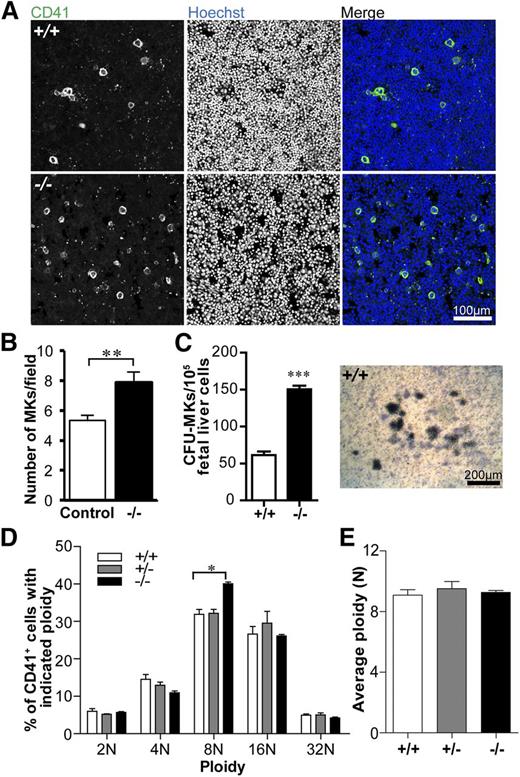
MK numbers are increased but ploidy is not significantly different in the absence of Tmod3
To investigate whether reduced platelet numbers in Tmod3−/− embryos are due to decreased MK number and/or impaired differentiation, we quantified CD41+ MKs in fetal liver cryosections. WT fetal livers contained sparsely distributed bright CD41+ MKs, as expected, whereas Tmod3−/− fetal livers contained ∼1.5-fold more CD41+ MKs (Figure 3A-B). This may reflect increased MK progenitors, based on increased colony-forming unit (CFU)-MKs in Tmod3−/− fetal livers (Figure 3C). We then examined the ploidy of isolated fetal liver MKs differentiated in vitro with mTPO (Figure 3D and supplemental Figure 2). As expected, the average ploidy of WT fetal liver MKs was close to ∼8N, which is lower than that of bone marrow MKs from adults (Figure 3D-E).48-50 However, the relative numbers of Tmod3−/− MKs with different ploidy were not significantly different, except for a small increase in 8N MKs (from 32% to 40%) (Figure 3D). Furthermore, there was no change in average ploidy (Figure 3E). Thus, alterations in MK numbers and ploidy are unlikely to account for decreased numbers of circulating platelets in Tmod3−/− embryos. Increased MKs and progenitors in Tmod3−/− fetal livers may reflect a compensatory response to the deficit in platelet production by Tmod3-deficient MKs (see below).
MK numbers are increased, whereas ploidy is not significantly changed in the absence of Tmod3. (A) Immunofluorescence staining for CD41 (green) and Hoechst (blue) on cryosections of Tmod3+/+ (top) and Tmod3−/− (bottom) fetal livers from E14.5 embryos. Images are single optical sections acquired using a Zeiss LSM 780 confocal laser scanning fluorescence microscope with a ×20 oil-immersion objective (N.A. 1.4). Bar, 100 µm. (B) Numbers of MKs per field (n = 46 fields from controls; 2 Tmod3+/+ and 2 Tmod3+/− fetal livers, and n = 42 fields from 2 Tmod3−/− fetal livers). **P < .05. (C) Numbers of CFU-MKs per 1 × 105 fetal liver cells from E15.5 embryos (n = 14 fields from 4 Tmod3+/+ fetal livers, n = 4 fields from 1 Tmod3−/− fetal liver) (left). Representative image of a WT CFU-MK colony (right). Tmod3−/− CFU-MK colonies were similar in appearance (not shown). Bar, 200 µm. ***P < .001 (D) Percentages of MKs with different ploidy (Tmod3+/+, n = 5; Tmod3+/−, n = 3; Tmod3−/−, n = 2 fetal livers). *P < .05. (E) Average MK ploidy from Tmod3+/+, Tmod3+/−, and Tmod3−/− fetal livers.
MK numbers are increased, whereas ploidy is not significantly changed in the absence of Tmod3. (A) Immunofluorescence staining for CD41 (green) and Hoechst (blue) on cryosections of Tmod3+/+ (top) and Tmod3−/− (bottom) fetal livers from E14.5 embryos. Images are single optical sections acquired using a Zeiss LSM 780 confocal laser scanning fluorescence microscope with a ×20 oil-immersion objective (N.A. 1.4). Bar, 100 µm. (B) Numbers of MKs per field (n = 46 fields from controls; 2 Tmod3+/+ and 2 Tmod3+/− fetal livers, and n = 42 fields from 2 Tmod3−/− fetal livers). **P < .05. (C) Numbers of CFU-MKs per 1 × 105 fetal liver cells from E15.5 embryos (n = 14 fields from 4 Tmod3+/+ fetal livers, n = 4 fields from 1 Tmod3−/− fetal liver) (left). Representative image of a WT CFU-MK colony (right). Tmod3−/− CFU-MK colonies were similar in appearance (not shown). Bar, 200 µm. ***P < .001 (D) Percentages of MKs with different ploidy (Tmod3+/+, n = 5; Tmod3+/−, n = 3; Tmod3−/−, n = 2 fetal livers). *P < .05. (E) Average MK ploidy from Tmod3+/+, Tmod3+/−, and Tmod3−/− fetal livers.
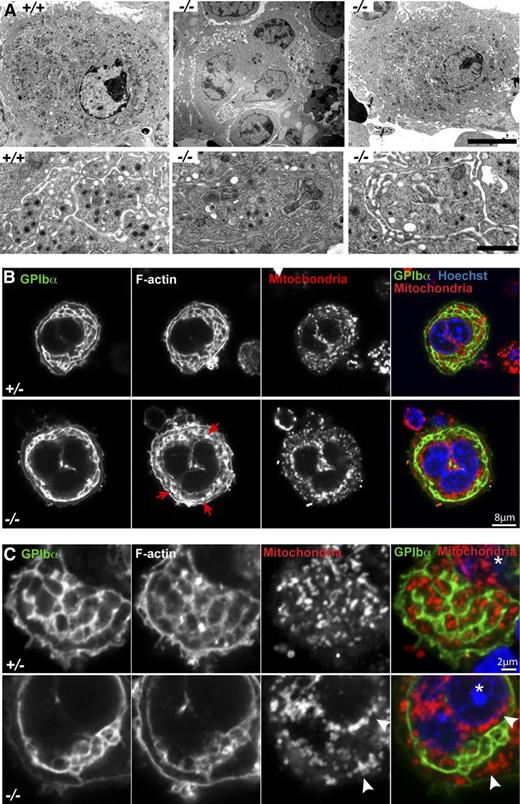
Tmod3−/− MKs exhibit abnormal DMS formation and organelle distributions
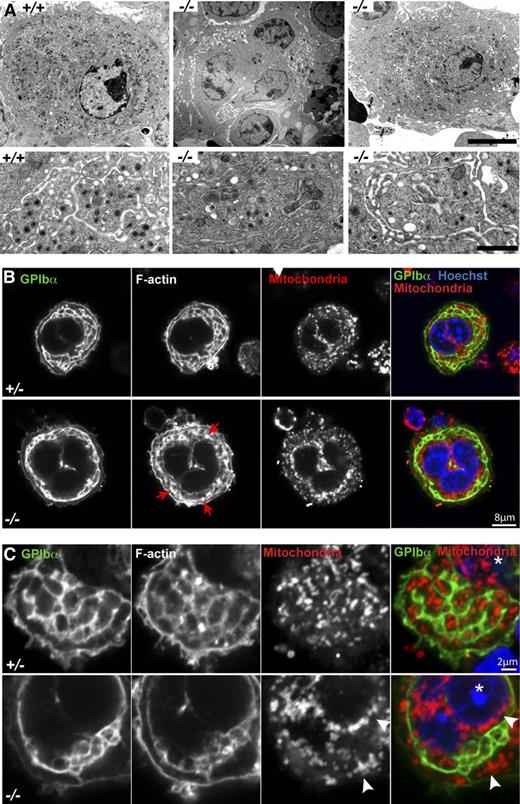
We next investigated whether the platelet deficits in Tmod3−/− embryos were due to abnormalities in MK cytoplasmic morphogenesis. TEM of mature MKs in WT fetal liver sections reveals an extensive DMS surrounding electron-dense granules within cytoplasmic platelet territories (Figure 4A, left). In contrast, Tmod3−/− MKs displayed two types of morphology, with about half resembling immature MKs containing large clumps of vesicles resembling a pre-DMS structure51 (Figure 4A, middle), and the other half containing relatively normal, elongated DMS tubules surrounding platelet territories (Figure 4A, right). In this latter type of Tmod3−/− MK, the DMS often enclosed larger cytoplasmic regions containing fewer electron-dense granules (Figure 4A, right).
Tmod3−/− MKs show abnormal ultrastructure with incomplete DMS formation, abnormal organelle distribution, and defects in F-actin. (A) Representative TEM images of Tmod3+/+ and Tmod3−/− MKs at lower (upper panels) and higher (lower panels) magnification. All Tmod3+/+ MKs showed DMS tubules surrounding platelet territories with abundant electron-dense α-granules (left). In contrast, about half of Tmod3−/− MKs contained large clumps of vesicles (middle), whereas others appeared with some DMS tubules surrounding platelet territories, which were often larger and with fewer electron-dense granules (right). Bar, 5 µm (upper panels) and 1 µm (lower panels). (B-C) Immunofluorescence staining of GPIbα, F-actin, mitochondria (TOM20), and nuclei (Hoechst) in isolated Tmod3+/− and Tmod3−/− fetal liver MKs. Grayscale images are shown for each stain, and the merge shows GPIbα (green), mitochondria (red), and nuclei (blue). In Tmod3+/− MKs, GPIbα and F-actin are associated with DMS membranes surrounding platelet territories that contain mitochondria. Tmod3+/− MK morphology is indistinguishable from Tmod3+/+ (data not shown). In Tmod3−/− MKs, GPIbα-labeled DMS membranes form fewer territories, which are deficient in mitochondria, F-actin accumulates in abnormal foci (B, red arrows), and mitochondria aggregate in the cytoplasm and around the nucleus (C, white arrowheads). Images are single optical sections selected from Z stacks acquired using a Zeiss LSM 780 confocal laser scanning fluorescence microscope with a ×100 oil-immersion objective (N.A. 1.4) at zoom 2 (B) or zoom 5 (C). Asterisk in (C), nucleus. Bars, 8 μm (B); 2 µm (C).
Tmod3−/− MKs show abnormal ultrastructure with incomplete DMS formation, abnormal organelle distribution, and defects in F-actin. (A) Representative TEM images of Tmod3+/+ and Tmod3−/− MKs at lower (upper panels) and higher (lower panels) magnification. All Tmod3+/+ MKs showed DMS tubules surrounding platelet territories with abundant electron-dense α-granules (left). In contrast, about half of Tmod3−/− MKs contained large clumps of vesicles (middle), whereas others appeared with some DMS tubules surrounding platelet territories, which were often larger and with fewer electron-dense granules (right). Bar, 5 µm (upper panels) and 1 µm (lower panels). (B-C) Immunofluorescence staining of GPIbα, F-actin, mitochondria (TOM20), and nuclei (Hoechst) in isolated Tmod3+/− and Tmod3−/− fetal liver MKs. Grayscale images are shown for each stain, and the merge shows GPIbα (green), mitochondria (red), and nuclei (blue). In Tmod3+/− MKs, GPIbα and F-actin are associated with DMS membranes surrounding platelet territories that contain mitochondria. Tmod3+/− MK morphology is indistinguishable from Tmod3+/+ (data not shown). In Tmod3−/− MKs, GPIbα-labeled DMS membranes form fewer territories, which are deficient in mitochondria, F-actin accumulates in abnormal foci (B, red arrows), and mitochondria aggregate in the cytoplasm and around the nucleus (C, white arrowheads). Images are single optical sections selected from Z stacks acquired using a Zeiss LSM 780 confocal laser scanning fluorescence microscope with a ×100 oil-immersion objective (N.A. 1.4) at zoom 2 (B) or zoom 5 (C). Asterisk in (C), nucleus. Bars, 8 μm (B); 2 µm (C).
To further investigate DMS morphology and organelle distribution in MKs, we immunostained the DMS with anti-GPIbα51,52 and mitochrondria with anti-TOM20, and imaged isolated MKs by confocal fluorescence microscopy (Figure 4B). Although all large-sized Tmod3+/+ or Tmod3+/− MKs with multilobular nuclei contained DMS membrane tubules organized into one or more rows of territories around the cell circumference, DMS tubule organization in Tmod3−/− MKs was variable, forming a narrow band of territories around the circumference of some MKs (Figure 4B), but only a few territories in others (Figure 4C). Strikingly, mitochondria were clearly visible within each platelet territory in Tmod3+/− MKs, but in Tmod3−/− MKs only some territories contained mitochondria, and many mitochondria collected in cytoplasmic aggregates near the nucleus (Figure 4B-C). Further, in yet other Tmod3−/− MKs, consistent with the TEM images (Figure 4A, middle), the DMS appeared as one or two large clumps of vesicles, resembling pre–DMS-like structures.51 Nonetheless, these Tmod3−/− MKs were large with similar multilobular nuclear morphology as Tmod3+/− MKs in which the DMS had formed tubules surrounding territories (supplemental Figure 3). Thus, although Tmod3−/− MKs achieve high nuclear ploidy similar to Tmod3+/+ and Tmod3+/− (Figure 3C-D), cytoplasmic morphogenesis is impaired, with abnormal DMS formation that does not create normal platelet territories with appropriate distributions of organelles.
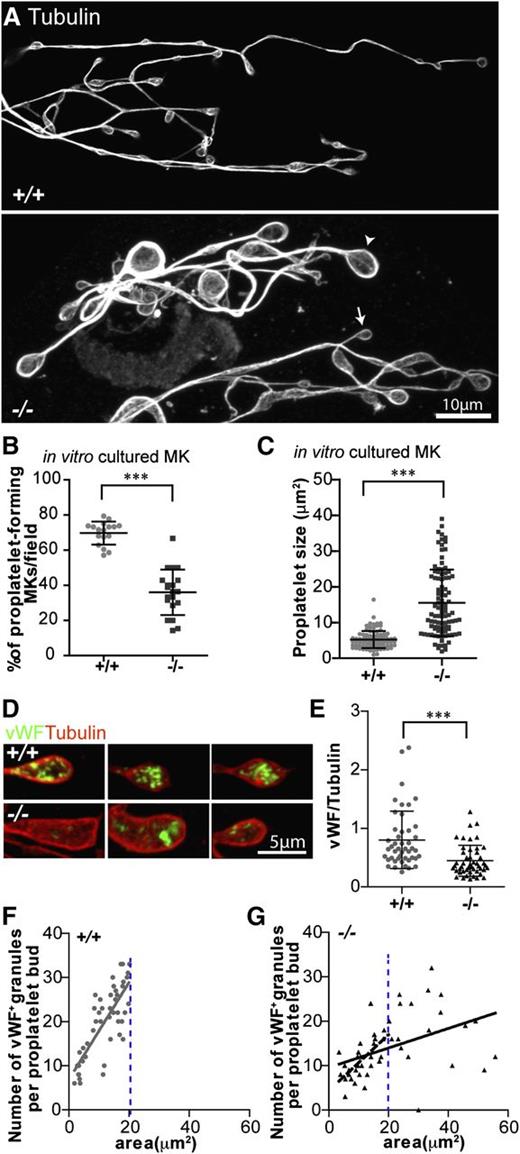
Proplatelet formation is impaired in Tmod3−/− MKs
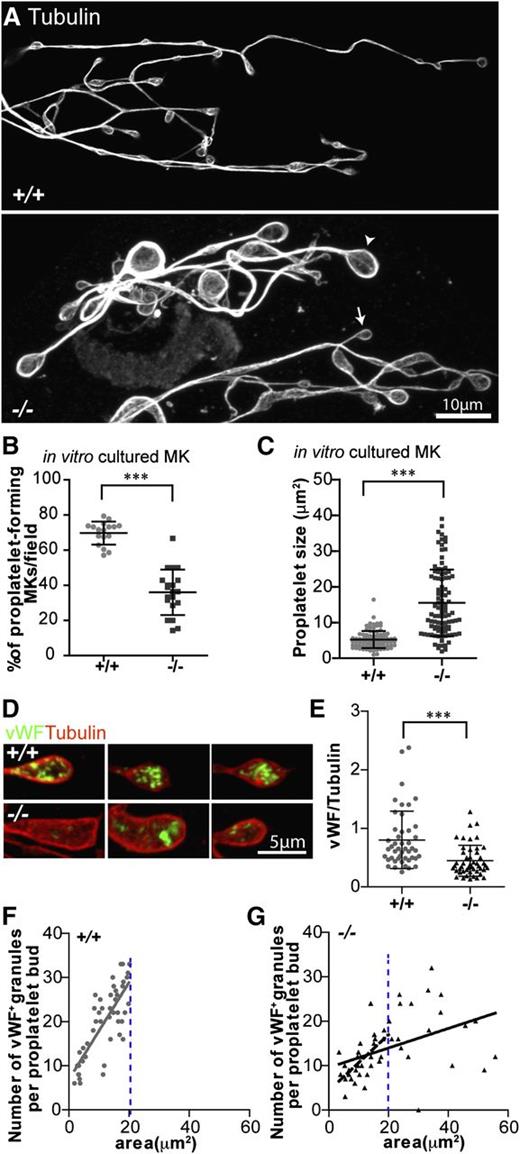
To test the ability of Tmod3−/− MKs to produce proplatelets, we cultured fetal liver MKs in mTPO and observed the morphologies and numbers of MKs forming proplatelets.43 Proplatelet buds formed from cultured Tmod3−/− MKs contained microtubule rings but were more variable in size and often much larger than WT proplatelets (Figure 5A). Furthermore, Tmod3−/− fetal livers contained about half as many proplatelet-forming MKs as WT fetal livers (Figure 5B), indicating that the proplatelet-forming ability of Tmod3−/− MKs is impaired. For the subset of Tmod3−/− MKs that did form proplatelets, proplatelet buds exhibited an approximately threefold increase in average area (Figure 5C), consistent with larger platelet sizes in embryonic blood (Figure 2E). The Tmod3−/− MKs that do not form proplatelets may correspond to MKs with clumped DMS vesicles (Figure 4A, middle, and supplemental Figure 3).
Proplaletet formation by Tmod3−/− MKs in culture is impaired, and Tmod3−/− proplatelets have abnormal levels of VWF+ granules. (A) Immunofluorescence staining of α-tubulin in proplatelets produced by MKs cultured in vitro from Tmod3+/+ and Tmod3−/− fetal livers reveals uniformly small sizes for Tmod3+/+, but highly variable sizes for Tmod3−/− proplatelets, including some very large ones. Arrowhead and arrow (lower panel) are examples of a large and small Tmod3−/− proplatelet, respectively. Bar, 10 µm. (B) Percentages of MKs cultured in vitro that form proplatelets. ***P < .001. (C) Sizes of proplatelet buds produced by cultured MKs. Tmod3+/+, 5.23 ± 2.39 µm2 (n = 96); Tmod3−/−, 15.48 ± 9.31 µm2 (n = 98). ***P < .001. Proplatelet sizes (areas, µm2) were measured from α-tubulin–stained proplatelets using Volocity 6.3 software to circle their perimeter. (D) Immunofluorescence staining of VWF (green) and α-tubulin (red) in proplatelet buds from cultured MKs. Bar, 5 µm. (E) Total VWF per proplatelet based on the ratio of VWF fluorescence intensity vs α-tubulin intensity. Tmod3+/+, 0.80 ± 0.49 (n = 46); Tmod3−/−, 0.45 ± 0.27 (n = 48). ***P < .001. (F-G) Number of VWF+-stained puncta per proplatelet bud with respect to area (solid grey line for Tmod3+/+, solid black line for Tmod3−/−). The number of VWF+ granules per proplatelet bud was linearly dependent on proplatelet area for Tmod3+/+ but not Tmod3−/− MKs (R2 = 0.6017 vs R2 = 0.2112, respectively). However, the subset of Tmod3−/− proplatelet buds with areas smaller than or equal to the area of the largest Tmod3+/+ proplatelet bud (demarcated by blue dotted line), revealed a stronger linear correlation (R2 = 0.4945, dashed black line). Thus, in Tmod3−/− proplatelets, the overall weakness of the linear relationship of the number of VWF+ granules per proplatelet bud as a function of proplatelet area is driven primarily by the unusually large, aberrant proplatelet buds. Images are single optical sections acquired using a Zeiss LSM 780 confocal laser scanning fluorescence microscope with a ×100 oil-immersion objective (N.A. 1.4) at zoom 0.6 (A) or zoom 2 (D).
Proplaletet formation by Tmod3−/− MKs in culture is impaired, and Tmod3−/− proplatelets have abnormal levels of VWF+ granules. (A) Immunofluorescence staining of α-tubulin in proplatelets produced by MKs cultured in vitro from Tmod3+/+ and Tmod3−/− fetal livers reveals uniformly small sizes for Tmod3+/+, but highly variable sizes for Tmod3−/− proplatelets, including some very large ones. Arrowhead and arrow (lower panel) are examples of a large and small Tmod3−/− proplatelet, respectively. Bar, 10 µm. (B) Percentages of MKs cultured in vitro that form proplatelets. ***P < .001. (C) Sizes of proplatelet buds produced by cultured MKs. Tmod3+/+, 5.23 ± 2.39 µm2 (n = 96); Tmod3−/−, 15.48 ± 9.31 µm2 (n = 98). ***P < .001. Proplatelet sizes (areas, µm2) were measured from α-tubulin–stained proplatelets using Volocity 6.3 software to circle their perimeter. (D) Immunofluorescence staining of VWF (green) and α-tubulin (red) in proplatelet buds from cultured MKs. Bar, 5 µm. (E) Total VWF per proplatelet based on the ratio of VWF fluorescence intensity vs α-tubulin intensity. Tmod3+/+, 0.80 ± 0.49 (n = 46); Tmod3−/−, 0.45 ± 0.27 (n = 48). ***P < .001. (F-G) Number of VWF+-stained puncta per proplatelet bud with respect to area (solid grey line for Tmod3+/+, solid black line for Tmod3−/−). The number of VWF+ granules per proplatelet bud was linearly dependent on proplatelet area for Tmod3+/+ but not Tmod3−/− MKs (R2 = 0.6017 vs R2 = 0.2112, respectively). However, the subset of Tmod3−/− proplatelet buds with areas smaller than or equal to the area of the largest Tmod3+/+ proplatelet bud (demarcated by blue dotted line), revealed a stronger linear correlation (R2 = 0.4945, dashed black line). Thus, in Tmod3−/− proplatelets, the overall weakness of the linear relationship of the number of VWF+ granules per proplatelet bud as a function of proplatelet area is driven primarily by the unusually large, aberrant proplatelet buds. Images are single optical sections acquired using a Zeiss LSM 780 confocal laser scanning fluorescence microscope with a ×100 oil-immersion objective (N.A. 1.4) at zoom 0.6 (A) or zoom 2 (D).
Proplatelet buds formed from cultured Tmod3−/− MKs had defective organelle composition, based on reduced staining for VWF, a marker of α-granules (Figure 5D-E). In proplatelet buds formed from Tmod3+/+ MKs, the number of VWF+ puncta is linearly related to proplatelet bud area (Figure 5F), as shown previously.10 However, in the absence of Tmod3, there are 2 populations of proplatelet buds: one containing VWF+ puncta in proportion to bud area, similar to WT, and another with abnormally large proplatelet buds, containing variable numbers of VWF+ puncta with no relationship to size (Figure 5G). The Tmod3−/− proplatelet buds with VWF+ content that scales with size may originate from regions of MK cytoplasm with relatively normal DMS tubule arrangements, whereas the abnormally large Tmod3−/− proplatelet buds with variable VWF+ content may originate from DMS-poor regions of MK cytoplasm with uneven organelle distributions (Figure 4).
F-actin polymerization is defective in Tmod3−/− MKs
To investigate whether defective F-actin polymerization underlies abnormalities in Tmod3−/− MKs and impaired proplatelet formation, we phalloidin-stained isolated fetal liver MKs. In control MKs, F-actin is associated with GPIbα-labeled DMS tubules and plasma membrane, with little cytoplasmic staining (Figure 4B-C). In contrast, in Tmod3−/− MKs, brightly stained cytoplasmic aggregates of F-actin are often present (Figure 4B, red arrows), in addition to F-actin at the DMS tubules and plasma membrane (Figure 4B-C). In the most abnormal Tmod3−/− MKs with no DMS-defined platelet territories, F-actin was present in the clumps of pre–DMS-like vesicles (supplemental Figure 3). Additionally, WT MKs extend protrusions and pseudopods with membrane-associated F-actin, whereas Tmod3−/− MKs form rudimentary protrusions, with cytoplasmic rather than membrane-associated F-actin (supplemental Figure 5).
To directly determine whether F-actin polymerization might be defective in Tmod3−/− MKs, we exploited the ability of MKs to assemble F-actin bundles upon spreading on collagen-coated coverslips. As expected, WT fetal liver-derived MKs assembled robust F-actin bundles as well as podosomes on their ventral surfaces (Figure 6A).11,17 Tmod3-stained puncta were often observed associated with the F-actin bundles, suggesting the F-actin pointed ends are capped (Figure 6A, bottom panels, arrowheads). NMIIA also assembles along the F-actin bundles of WT MKs (Figure 6B), appearing in a striated pattern indicative of their contractile function.53 In contrast, in Tmod3−/− MKs spreading on collagen, F-actin polymerization with NMIIA into bundles is severely impaired (Figure 6C). Instead, F-actin forms amorphous aggregates near the periphery of the spreading MKs, and NMIIA does not assemble (Figure 6C). Thus, Tmod3 regulates F-actin polymerization in fetal liver-derived MKs.
Tmod3 regulates F-actin polymerization and organization in MKs. (A) Tmod3 is associated with F-actin bundles that assemble in MKs spread on collagen. Immunofluorescence staining of F-actin (red) and Tmod3 (green) in WT MKs spread on collagen for 30 minutes. F-actin polymerizes into bundles and foci on the ventral surface of the MKs. Boxed regions shown at high magnification (lower panels) reveal Tmod3 puncta associated with the F-actin bundles (yellow arrrowheads). Bars, 10 μm. (B-C) Immunofluorescence staining of F-actin (red) and NMIIA (green) in Tmod3+/+ and Tmod3−/− MKs spread on collagen. In WT MKs, F-actin assembles into parallel bundles, as well as intensely stained foci on the ventral surface, termed podosomes.17 NMIIA is associated with the F-actin bundles (but not the podosomes) in a striated pattern in WT MKs, whereas both F-actin and NMIIA form amorphous aggregates near the cell periphery in Tmod3−/− MKs. Boxed regions shown at higher magnification (lower sets of panels in [B-C]). Scale bars, 10 µm. Images are single optical sections from the ventral surface of MKs, acquired using a Zeiss LSM 780 confocal laser scanning fluorescence microscope with a ×63 oil-immersion objective (N.A. 1.4) at zoom 1 (top panels) and zoom 3 (boxed regions).
Tmod3 regulates F-actin polymerization and organization in MKs. (A) Tmod3 is associated with F-actin bundles that assemble in MKs spread on collagen. Immunofluorescence staining of F-actin (red) and Tmod3 (green) in WT MKs spread on collagen for 30 minutes. F-actin polymerizes into bundles and foci on the ventral surface of the MKs. Boxed regions shown at high magnification (lower panels) reveal Tmod3 puncta associated with the F-actin bundles (yellow arrrowheads). Bars, 10 μm. (B-C) Immunofluorescence staining of F-actin (red) and NMIIA (green) in Tmod3+/+ and Tmod3−/− MKs spread on collagen. In WT MKs, F-actin assembles into parallel bundles, as well as intensely stained foci on the ventral surface, termed podosomes.17 NMIIA is associated with the F-actin bundles (but not the podosomes) in a striated pattern in WT MKs, whereas both F-actin and NMIIA form amorphous aggregates near the cell periphery in Tmod3−/− MKs. Boxed regions shown at higher magnification (lower sets of panels in [B-C]). Scale bars, 10 µm. Images are single optical sections from the ventral surface of MKs, acquired using a Zeiss LSM 780 confocal laser scanning fluorescence microscope with a ×63 oil-immersion objective (N.A. 1.4) at zoom 1 (top panels) and zoom 3 (boxed regions).
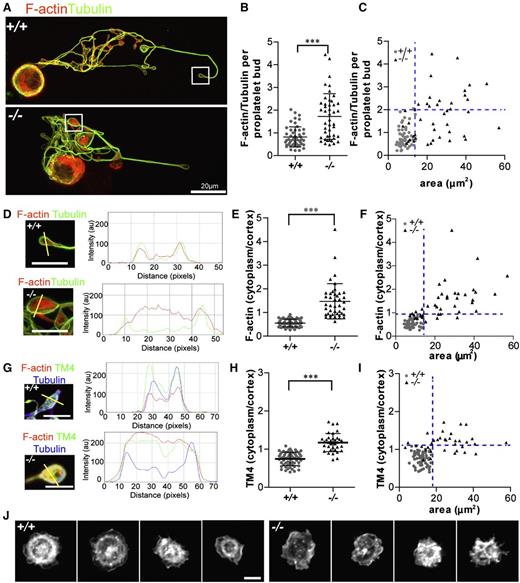
F-actin and TM4 organization are perturbed in Tmod3−/− proplatelet buds
Tmod3 binds TMs to cap and stabilize F-actin pointed ends,33 with TM4 as the predominant TM in platelets.42,54-56 Indeed, TM4 is detected in whole-cell lysates of in vitro cultured MKs from mouse fetal livers (supplemental Figure 4A-B), whereas long, muscle-type TMs are not detected (data not shown). Co-IP shows that MK Tmod3 and TM4 are associated with actin and associated cytoskeletal proteins (NMIIA, FLNA, and β2-spectrin) (supplemental Figure 4A-B). Confocal fluorescence microscopy of proplatelets further reveals that Tmod3 and TM4 partially colocalize with F-actin and are enriched at the cortex of proplatelet buds (supplemental Figure 4C-F), similar to cortical enrichment of F-actin in circulating embryonic platelets (supplemental Figure 1).
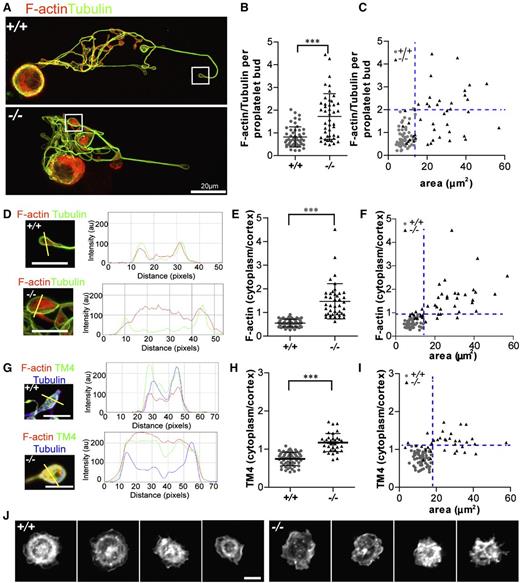
We hypothesized that absence of Tmod3 leading to disruption of Tmod3–TM4-actin associations might alter F-actin abundance and/or organization in Tmod3−/− proplatelet buds. Tmod3−/− proplatelet buds exhibited highly variable F-actin levels, with some similar to WT and others with up to approximately four- to fivefold more F-actin (Figure 7A-B). Moreover, similar to VWF+ puncta (Figure 5G), normal-sized Tmod3−/− proplatelet buds had relatively normal F-actin levels, whereas larger buds had highly variable levels of F-actin (Figure 7C).
Tmod3−/− proplatelets have defects in F-actin and TM4 organization, and circulating Tmod3−/− platelets fail to organize an F-actin ring after spreading on collagen. (A) Immunofluorescence staining of F-actin (red) and α-tubulin (green) in proplatelet-forming Tmod3+/+ (top) and Tmod3−/− (bottom) fetal liver MKs. Bar, 20 µm. (B) Relative level of F-actin vs α-tubulin per proplatelet bud, calculated from fluorescence intensity ratios as in Figure 5. Tmod3+/+: 0.82 ± 0.44 (n = 51); Tmod3−/−: 1.72 ± 1.0 (n = 45). ***P < .001. (C) F-actin/tubulin intensity per proplatelet bud with respect to area. (D) High magnification images and representative line scans of α-tubulin (green) and F-actin (red) fluorescence in Tmod3+/+ and Tmod3−/− proplatelets. Bars, 5 µm. (E) F-actin cytoplasm vs cortex intensity per proplatelet bud. Tmod3+/+: 0.55 ± 0.17 (n = 50); Tmod3−/−: 1.47 ± 0.74 (n = 42). ***P < .001. (F) F-actin cytoplasm vs cortex intensity per proplatelet bud with respect to area. (G) High magnification images and representative line scans of TM4 (green), α-tubulin (blue), and F-actin (red) fluorescence in Tmod3+/+ and Tmod3−/− proplatelets. Bars, 5 µm. (H) TM4 cytoplasm vs cortex intensity per proplatelet bud. Tmod3+/+: 0.75 ± 0.17 (n = 60); Tmod3−/−: 1.17 ± 0.24 (n = 60). (I) TM4 cytoplasm vs cortex intensity per proplatelet bud with respect to area. (J) Rhodamine-phalloidin staining for F-actin in Tmod3+/+ (left) and Tmod3−/− (right) embryonic platelets spread on collagen for 30 minutes. Bar, 2 µm. Images are single optical sections acquired using a Zeiss LSM 780 confocal laser scanning fluorescence microscope with a 100× oil-immersion objective (N.A. 1.4) at zoom 1 (A) or zoom 3 (D,G,J). Fluorescence intensities of α-tubulin and F-actin in proplatelets (B-C) were determined using ImageJ software, with areas of proplatetets determined from α-tubulin staining as described in Figure 5. Ratios of cytoplasm to cortex staining intensities for F-actin and TM4 (E-F,H-I) were determined from intensities within small ∼0.5 μm diameter circles placed over the edge or middle of the proplatelet, using ImageJ software. Line scans of TM4 and F-actin signals across proplatelets were performed also with ImageJ software (D,G). Blue dotted lines indicate maximum x- and y-coordinate values of Tmod3+/+ data.
Tmod3−/− proplatelets have defects in F-actin and TM4 organization, and circulating Tmod3−/− platelets fail to organize an F-actin ring after spreading on collagen. (A) Immunofluorescence staining of F-actin (red) and α-tubulin (green) in proplatelet-forming Tmod3+/+ (top) and Tmod3−/− (bottom) fetal liver MKs. Bar, 20 µm. (B) Relative level of F-actin vs α-tubulin per proplatelet bud, calculated from fluorescence intensity ratios as in Figure 5. Tmod3+/+: 0.82 ± 0.44 (n = 51); Tmod3−/−: 1.72 ± 1.0 (n = 45). ***P < .001. (C) F-actin/tubulin intensity per proplatelet bud with respect to area. (D) High magnification images and representative line scans of α-tubulin (green) and F-actin (red) fluorescence in Tmod3+/+ and Tmod3−/− proplatelets. Bars, 5 µm. (E) F-actin cytoplasm vs cortex intensity per proplatelet bud. Tmod3+/+: 0.55 ± 0.17 (n = 50); Tmod3−/−: 1.47 ± 0.74 (n = 42). ***P < .001. (F) F-actin cytoplasm vs cortex intensity per proplatelet bud with respect to area. (G) High magnification images and representative line scans of TM4 (green), α-tubulin (blue), and F-actin (red) fluorescence in Tmod3+/+ and Tmod3−/− proplatelets. Bars, 5 µm. (H) TM4 cytoplasm vs cortex intensity per proplatelet bud. Tmod3+/+: 0.75 ± 0.17 (n = 60); Tmod3−/−: 1.17 ± 0.24 (n = 60). (I) TM4 cytoplasm vs cortex intensity per proplatelet bud with respect to area. (J) Rhodamine-phalloidin staining for F-actin in Tmod3+/+ (left) and Tmod3−/− (right) embryonic platelets spread on collagen for 30 minutes. Bar, 2 µm. Images are single optical sections acquired using a Zeiss LSM 780 confocal laser scanning fluorescence microscope with a 100× oil-immersion objective (N.A. 1.4) at zoom 1 (A) or zoom 3 (D,G,J). Fluorescence intensities of α-tubulin and F-actin in proplatelets (B-C) were determined using ImageJ software, with areas of proplatetets determined from α-tubulin staining as described in Figure 5. Ratios of cytoplasm to cortex staining intensities for F-actin and TM4 (E-F,H-I) were determined from intensities within small ∼0.5 μm diameter circles placed over the edge or middle of the proplatelet, using ImageJ software. Line scans of TM4 and F-actin signals across proplatelets were performed also with ImageJ software (D,G). Blue dotted lines indicate maximum x- and y-coordinate values of Tmod3+/+ data.
Next, we evaluated F-actin and TM4 distributions in proplatelet buds. Tmod3+/+ proplatelet buds exhibited cortical enrichment of F-actin and TM4 (Figure 7D,G, supplemental Figure 4D,F, and supplemental Figure 6). However, in Tmod3−/− proplatelet buds, the relative F-actin and TM4 intensity in the cytoplasm was increased as compared with the cortex, indicating TM4 and F-actin redistribution (Figure 7D,G and supplemental Figure 6). Quantification showed an approximate threefold increase for F-actin in the cytoplasmic vs cortical intensity ratio per proplatelet bud (Figure 7E), and an approximate twofold increase for TM4 (Figure 7H). Notably, the increased cytoplasm/cortex ratios of F-actin and TM4 were more pronounced in the abnormally large Tmod3−/− proplatelet buds (Figure 7F,I). Thus, the deletion of Tmod3 disrupts TM4–F-actin cytoskeletal organization, with more pronounced effects in abnormally large proplatelet buds.
Abnormal F-actin organization was also evident in Tmod3−/− circulating embryonic platelets, where foci and F-actin aggregates were present in the platelet interior, in contrast to Tmod3+/+ embryonic platelets where F-actin concentrates at the cortex were present (supplemental Figure 1). We then examined F-actin organization in embryonic platelets spreading on collagen-coated coverslips. WT platelets formed peripheral lamellipodia with F-actin at the leading edge, as well as an internal F-actin ring and central concentration (Figure 7J). In contrast, Tmod3−/− platelets formed lamellipodia at their periphery, but not the internal F-actin ring, and, in others, F-actin formed irregular bundles or partial rings (Figure 7J). Thus, Tmod3 regulates F-actin organization in proplatelets from cultured MKs and in embryonic circulating platelets.
Discussion
We identified a new feature of Tmod3−/− mouse embryos: hemorrhaging with macrothrombocytopenia. Tmod3 is the only Tmod in mouse fetal liver MKs and platelets, and is important for cytoplasmic morphogenesis of MKs. Tmod3−/− MKs display an immature or incomplete DMS, which forms insufficient tubules for platelet territories and the membrane reservoir required for proplatelet protrusion, leading to impaired ability to produce proplatelets. Tmod3−/− MKs may contain pre–DMS-like vesicular clumps,51 due to delay or inability to remodel the DMS, or contain reduced DMS tubules creating fewer platelet territories with irregular organelle distributions. Tmod3−/− MKs produce 2 varieties of proplatelet buds: ones within a normal size range (∼2 to 20 μm2),10 and others that are abnormally large (up to ∼60 μm2). In normal-sized WT and Tmod3−/− proplatelet buds, VWF+ granule content scales with proplatelet bud size, suggesting that Tmod3 regulation of F-actin is not required for organelle transport down proplatelet extensions, which depends on microtubule-dependent transport and sliding processes.8,57 However, F-actin polymerization is aberrant in Tmod3−/− MKs, and increased F-actin and redistribution of F-actin and TM4 from the cortex to the cytoplasm is only observed in abnormally large Tmod3−/− proplatelet buds. Therefore, normal-size Tmod3−/− proplatelet buds may originate from regions of MK cytoplasm containing DMS tubules, whereas abnormally large proplatelet buds may originate from DMS-poor regions of cytoplasm with irregular distributions of organelles and F-actin aggregates.
How might Tmod3 control MK DMS formation, organelle distribution, and proplatelet formation? The DMS is associated with F-actin (Figure 4B-C)23 in a spectrin-actin network,24 and spectrin tetramer formation and F-actin assembly are known to influence DMS formation and platelet biogenesis.14,23,24,58,59 Spectrin is also implicated in intracellular vesicular transport,60-64 suggesting Tmod3 may influence DMS and organelle distributions in MKs via stabilization of spectrin-actin networks associated with intracellular membranes. A precedent for direct association of Tmod3 with an intracellular membrane receptor is that of Tmod3 with sAnk1.5, a transmembrane protein in the sarcoplasmic reticulum of skeletal muscle.36 Alternatively, Tmod3 may associate with MK and proplatelet membranes indirectly via F-actin’s links to spectrin networks or to FLNA, which binds GP1bα.7,25
Several lines of evidence in our study support the idea that impaired DMS formation and aberrant organelle distributions in Tmod3−/− MKs are caused by a disrupted F-actin cytoskeleton. Tmod3−/− MKs display aberrant cytoplasmic accumulations of F-actin (Figure 4B, supplemental Figures 3 and 5), and fail to polymerize F-actin during spreading on collagen (Figure 6C). Tmod3 is present in these F-actin bundles in WT MKs (Figure 6A), consistent with Tmod3 capping and promotion of F-actin assembly in the MK actin cytoskeleton. In vitro, Tmod3’s F-actin–capping activity is enhanced by Tmod3 binding TM, which binds along F-actin and stabilizes filaments.26,30,33 TM4 (TPM4 gene) is the primary TM in platelets42,54-56 and MKs (Figure 1 and supplemental Figure 4), and Tmod3 associates with TM4 and the actin cytoskeleton, which includes FLNA, β2-spectrin, and NMIIA (supplemental Figure 4). Since Tmod3 binds directly to TMs,30,33 including TM4,65 it is likely that Tmod3 caps TM4-coated F-actin linked to FLNA, β2-spectrin, and/or NMIIA in MKs and proplatelets.
Loss of Tmod3 leads to F-actin instability,30,33,34 yet overall, more F-actin is observed in large Tmod3−/− proplatelet buds (Figure 7B,C), and F-actin aggregates are observed in Tmod3−/− MK cytoplasm (Figure 4B and supplemental Figure 5). What might explain this? Increased actin monomer levels resulting from disassembly of Tmod3-capped F-actin would be available for polymerization nucleated by other actin assembly factors present in MKs and proplatelets, such as WASp17,58 or mDia1.20 In other cell types, assembly of diverse F-actin structures are promoted by actin nucleation factors that compete for a limited pool of globular actin; thus when one assembly factor and pathway is eliminated, the others promote excessive F-actin assembly into alternative cytoskeletal structures.66,67
The actin cytoskeletal proteins that play important roles in MK platelet biogenesis (FLNA, ADF/n-cofilin, Rac/Cdc42, RhoA, and WASp) are also required for platelet spreading, adhesion, and granule secretion to form clots.7,68,69 Indeed, Tmod3-null embryonic platelets spreading on collagen display defective F-actin polymerization and reorganization (Figure 7J). Although Tmod3-null platelets form lamellipodia, an actin–polymerization-dependent process, they do not form the internal F-actin rings observed during organelle centralization.47,68 Moreover, abnormal numbers of VWF+ granules in Tmod3−/− proplatelets (Figure 5), and fewer granules and more empty vesicular structures observed by TEM in circulating platelets in Tmod3−/− embryos (Figure 2) support the idea that Tmod3−/− platelet function is likely defective. Future studies with a MK-restricted Tmod3 deletion in adult mice will be required to define the precise role of Tmod3 in platelet function in thrombosis and hemostasis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Malcolm Wood in the The Scripps Research Institute Core Microscopy Facility for TEM of MKs and platelets, Zaverio Ruggeri and Alessandro Zarpellon for anti-GPIbα antibodies and discussions, and David Gokhin for help with editing.
This study was supported by grants from the National Institutes of Health, National Heart, Lung, and Blood Institute (R01-HL083464) (V.M.F), and National Institute of Diabetes and Digestive and Kidney Diseases (R01 DK094934 and R01 DK086267) (D.S.K.), and funding from the State of Connecticut Stem Cell Research Fund (D.S.K).
Authorship
Contribution: Z.S. analyzed protein expression, performed MK cultures, confocal microscopy and quantitative image analyses, prepared figures, and wrote portions of the manuscript; R.B.N. bred mice, performed embryo analysis, histology and confocal microscopy, and prepared the figures; C.S. and S.H. performed fetal liver dissections for flow cytometry, analyzed data, and prepared the figures; D.S.K. provided intellectual input, experimental design, data analysis, and read the manuscript; and V.M.F. conceived the project, designed experiments, interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Velia M. Fowler, Department of Cell and Molecular Biology, CB163, The Scripps Research Institute, 10550 N. Torrey Pines Rd, La Jolla, CA 92037; e-mail: velia@scripps.edu.
References
Author notes
Z.S. and R.B.N. contributed equally to this study.

![Figure 6. Tmod3 regulates F-actin polymerization and organization in MKs. (A) Tmod3 is associated with F-actin bundles that assemble in MKs spread on collagen. Immunofluorescence staining of F-actin (red) and Tmod3 (green) in WT MKs spread on collagen for 30 minutes. F-actin polymerizes into bundles and foci on the ventral surface of the MKs. Boxed regions shown at high magnification (lower panels) reveal Tmod3 puncta associated with the F-actin bundles (yellow arrrowheads). Bars, 10 μm. (B-C) Immunofluorescence staining of F-actin (red) and NMIIA (green) in Tmod3+/+ and Tmod3−/− MKs spread on collagen. In WT MKs, F-actin assembles into parallel bundles, as well as intensely stained foci on the ventral surface, termed podosomes.17 NMIIA is associated with the F-actin bundles (but not the podosomes) in a striated pattern in WT MKs, whereas both F-actin and NMIIA form amorphous aggregates near the cell periphery in Tmod3−/− MKs. Boxed regions shown at higher magnification (lower sets of panels in [B-C]). Scale bars, 10 µm. Images are single optical sections from the ventral surface of MKs, acquired using a Zeiss LSM 780 confocal laser scanning fluorescence microscope with a ×63 oil-immersion objective (N.A. 1.4) at zoom 1 (top panels) and zoom 3 (boxed regions).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/126/4/10.1182_blood-2014-09-601484/4/m_520f6.jpeg?Expires=1769315888&Signature=b2z3XW4artFFFpYZYI4ocNhWOKDHST2iKaUJUT1iqlRs6KOx0C8KLnMFLxsdZUVG2Z8vBaauua9gt7U-TZF66cz5lVz6B~IvNIMZFvdCGXMmEKDAP4a5vhi3OIEb7FTQnfVVUJoO6xhFNTqdceCEtyFcPBByNtXlGHgl-bpWglOnD0jZSb8z8ZsZnhOG5Y4u6uAOnULDXnNW-yFR7HBVENk8VLIwoMGdXQPPFKnvSXNJcbK8SyHlHjA0KLrDMSOb99xCOwd8GOoNlS67O5FNFARjGKkXdUeERyHBL4Fky~KZYmN1ZaY0PR2IiDCg5mAbLXOdG22~LYOez0jWztn6Yw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 6. Tmod3 regulates F-actin polymerization and organization in MKs. (A) Tmod3 is associated with F-actin bundles that assemble in MKs spread on collagen. Immunofluorescence staining of F-actin (red) and Tmod3 (green) in WT MKs spread on collagen for 30 minutes. F-actin polymerizes into bundles and foci on the ventral surface of the MKs. Boxed regions shown at high magnification (lower panels) reveal Tmod3 puncta associated with the F-actin bundles (yellow arrrowheads). Bars, 10 μm. (B-C) Immunofluorescence staining of F-actin (red) and NMIIA (green) in Tmod3+/+ and Tmod3−/− MKs spread on collagen. In WT MKs, F-actin assembles into parallel bundles, as well as intensely stained foci on the ventral surface, termed podosomes.17 NMIIA is associated with the F-actin bundles (but not the podosomes) in a striated pattern in WT MKs, whereas both F-actin and NMIIA form amorphous aggregates near the cell periphery in Tmod3−/− MKs. Boxed regions shown at higher magnification (lower sets of panels in [B-C]). Scale bars, 10 µm. Images are single optical sections from the ventral surface of MKs, acquired using a Zeiss LSM 780 confocal laser scanning fluorescence microscope with a ×63 oil-immersion objective (N.A. 1.4) at zoom 1 (top panels) and zoom 3 (boxed regions).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/126/4/10.1182_blood-2014-09-601484/4/m_520f6.jpeg?Expires=1769315889&Signature=PevZ0XjnjSNFSDOHP2lqXFx4Mhs9KGjlH~iue1eCUt0jTpd-WjtVMt7nzpZLMMkfyJQyX~nRKhEA7yEpjL9zugmByzyuwismcD1iQxsin8z0Z3eXcgwgSotZnUVTvRd6Kxmow9cy2ZfpvWtHZNktd1wD~XSsN3krZGAlalm7zVGzxPgAKd4ULLT9EuPs10BJun6cfqLinip~iIniV6tjxiyiMY~W9y99kRii-LjXmowYsxpER7BxNIMu7abfReKPaxwYygpwqZEBOR1jSoN-UDZ9D8bnZ2SOM72uB0pmn4ez9nMeYbu0x7K4G1-hqkmNYSZwS7QTfoHgk5GSJJuGsA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)