Key Points
FXIIIa exhibits a preference for Q237 in crosslinking reactions within fibrinogen αC (233-425) followed by Q328 and Q366.
None of the reactive glutamines in αC 233-425 (Q237, Q328, and Q366) are required to react first before the others can crosslink.
Abstract
Factor XIIIa (FXIIIa) introduces covalent γ-glutamyl-ε-lysyl crosslinks into the blood clot network. These crosslinks involve both the γ and α chains of fibrin. The C-terminal portion of the fibrin α chain extends into the αC region (210-610). Crosslinks within this region help generate a stiffer clot, which is more resistant to fibrinolysis. Fibrinogen αC (233-425) contains a binding site for FXIIIa and three glutamines Q237, Q328, and Q366 that each participate in physiological crosslinking reactions. Although these glutamines were previously identified, their reactivities toward FXIIIa have not been ranked. Matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) mass spectrometry and nuclear magnetic resonance (NMR) methods were thus used to directly characterize these three glutamines and probe for sources of FXIIIa substrate specificity. Glycine ethyl ester (GEE) and ammonium chloride served as replacements for lysine. Mass spectrometry and 2D heteronuclear single quantum coherence NMR revealed that Q237 is rapidly crosslinked first by FXIIIa followed by Q366 and Q328. Both 15NH4Cl and 15N-GEE could be crosslinked to the three glutamines in αC (233-425) with a similar order of reactivity as observed with the MALDI-TOF mass spectrometry assay. NMR studies using the single αC mutants Q237N, Q328N, and Q366N demonstrated that no glutamine is dependent on another to react first in the series. Moreover, the remaining two glutamines of each mutant were both still reactive. Further characterization of Q237, Q328, and Q366 is important because they are located in a fibrinogen region susceptible to physiological truncations and mutation. The current results suggest that these glutamines play distinct roles in fibrin crosslinking and clot architecture.
Introduction
Abnormal fibrinogen levels and fibrin clot structure have been associated with several pathological conditions including cardiovascular disease, arteriosclerosis, and bleeding disorders.1,2 The N-termini of the fibrinogen (AαBβγ)2 chains form the central E region, which extends via a coil-coiled region to two terminal D regions. A flexible portion of the Aα chain, the αC region, extends from the ends of the D regions and is tethered to the central E region.3,4 To initiate fibrin polymerization, thrombin cleaves fibrinopeptides A and B from the Aα and Bβ chains of fibrinogen to form fibrin monomers (αβγ)2.5,6 These cleavages lead to protofibril formation among the monomers and also releases the αC region.4,7,8
At the final stage of blood coagulation, factor XIIIa (FXIIIa) introduces γ-glutamyl-ε-lysyl isopeptide bonds between selective glutamines (Q) and lysines (K) within fibrin to form a clot with enhanced elastic properties.6,9-13 Moreover, the overall fibrin network has decreased fiber diameter, increased fiber density, reports of thinner fibers, and increased clot stiffness.11,14-16 FXIIIa first crosslinks fibrin γQ398 or γQ399 to fibrin γK406 thereby forming γ-γ dimers. Crosslinks involving the fibrin α chain appears later and involves the generation of α-α dimers, γ-α hybrids, and higher α-α polymers.17-20 Five known reactive glutamines in the αC region include Q221 (and/or 223), Q237, Q328, and Q366.16,21-23 FXIIIa also crosslinks α2-antiplasmin (α2AP), plasmin activator inhibitor 2 (PAI-2), and fibronectin into the fibrin clot network.24-27 FXIII(a) and fibrinogen play additional supporting roles in the presence of red blood cells. FXIII binding to fibrinogen γ390-396 and subsequent crosslinking of the α chain helps mediate red blood cell retention in venous thrombi.28,29
The αC region of fibrinogen contributes its own critical roles in the assembly and properties of clot structure.8,30-33 This region has also been directly correlated with adhesion events.34-36 Fibrinogen αC region (221-610) is composed of a flexible αC connector (221-391) and a more structured αC domain (392-610).4,7 During fibrin polymerization, crosslinking in the αC region has been shown to promote lateral aggregation and protofibril staggering.8,30,37 Studies with native fibrinogen vs the mutant (γQ398N, γQ399N, and γK406R) have revealed independent contributions coming from α-α crosslinks.16,38 Such crosslinks play major roles in promoting clot stiffness, fiber straightening, and hindering fibrinolysis.
Truncations to the αC region have been shown to alter the nature of the clot formed.32 With fibrinogen Otago, the Aα chain ends at P270, resulting in severe hypofibrinogenemia. Further analysis revealed that loss of (271-610) leads to impaired fibrin polymerization and the production of much thicker fibers.32,33,39 Fibrinogen Seoul II (Q328P mutation) exhibits impaired fibrin polymerization and a reduced extent of α-α crosslinking due in part to the loss of a reactive glutamine within the αC region.40
Several studies have identified specific crosslinking sites in the αC region responsible for lateral aggregation, but little is known about the individual reactive glutamines and the roles played by the surrounding residues.17,18,20,22,23,41,42 Antibody studies by Procyk et al identified a FXIIIa binding site within αC (242-424) and then narrowed the site more specifically to αC (389-402).43 Using recombinant αC (233-425), Smith et al demonstrated that the key contact region for FXIII A2′ (thrombin-activated) and A2B2 involves αC (371-425) with E396 playing an important role.44,45 In close proximity to this FXIII binding region, αC (233-425) contains three reactive glutamines (Q237, Q328, and Q366) that each become crosslinked to a selective set of lysines within the fibrin clot network.17,23
The aim of the current study was to characterize the glutamines within the fibrinogen αC region (233-425) that are known to be crosslinked by FXIIIa under physiological conditions. Matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) mass spectrometry and 2D heteronuclear single quantum coherence (HSQC) nuclear magnetic resonance (NMR) assays were used to directly monitor FXIIIa-catalyzed reactions between the substrate glutamines in αC (233-425) and a set of lysine mimics.46 The results demonstrate that Q237 is the most reactive glutamine of fibrinogen αC (233-425), followed by Q366 and Q328. Moreover, no glutamine is dependent on another to react first in the series. New knowledge gained about these three glutamines is considered relative to the conformational features of fibrinogen, and to the mutants fibrinogen Otago and fibrinogen Seoul II.
Materials and methods
Proteins and chemicals
Human cellular FXIII was a generous gift from the late Dr Paul Bishop. Stock aliquots of FXIII were prepared in 18 MΩ deionized water and the concentration measured on a Cary 100 Bio UV-Visible Spectrophotometer using an extinction coefficient of 1.49 (mL mg−1 cm−1) and stored at −70°C. Thrombin and glycine ethyl ester (GEE) were obtained from Sigma-Aldrich (St. Louis, MO). 15NH4Cl and 15N-GEE were obtained from Cambridge Isotope Laboratories (Andover, MA).
Site-directed mutagenesis of αC (233-425) to generate Q to N substitutions
The pGEX-6P-1 plasmid vector contained complementary DNA for glutathione S-transferase (GST)-tagged αC fragment (233-425). This αC sequence was derived from the full-length fibrinogen Aα chain complementary DNA.44 Individual glutamine substitutions, Q237N, Q328N, and Q366N were introduced into αC (233-425) using the QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA). The resulting substitutions were confirmed with DNA sequencing. DNAs coding for the different αC (233-425) species were transformed into BL21 Gold DE3 Escherichia coli cells.
Fibrinogen αC (233-425) expression
GST-αC (233-425) and its mutants were expressed as described previously by Smith et al,44 with few modifications. Briefly, GST-αC expressing cells were incubated in Terrific Broth (1.2% tryptone, 2.4% yeast extract, 0.4% glycerol, 742 mM K2HPO4, 171 mM KH2PO4, and 0.1 mg/mL ampicillin) at 37°C on a rotary shaker until the optical density at 600 nm reached 0.9. αC expression was induced with 1 mM isopropyl β-D-1-thiogalactopyranoside followed by incubation at 30°C for 16 hours. Cells were harvested by centrifugation at 5000 g at 4°C. Biomass was resuspended in Wash Buffer (20 mM Tris Base, pH 8.0, 50 mM NaCl) and centrifuged at 5000 g at 4°C. Resulting pellets were stored at −20°C.
Frozen pellets were resuspended in phosphate buffer saline: 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) and incubated with 1 mM dithiothreitol and 1 mg/mL lysozyme, followed by the addition of 2 μg/mL aprotinin, 1 μM pepstatin A, 10 μM leupeptin, 4 mM benzamidine hydrochloride, 0.5 mM phenylmethylsulfonyl fluoride, 0.05% sodium deoxycholate, 0.02% triton X-100, 5 μg/mL Dnase I, and 5 mM MgCl2. The Dnase I reaction was quenched with 6 mM EDTA and the lysate was centrifuged at 22 000 g at 4°C. Supernatant was supplemented with 1% (w/v) streptomycin sulfate and centrifuged at 22 000 g at 4°C. The resulting soluble fraction was passed through a 0.2 μm membrane filter and loaded on a GST-affinity column using an AKTAprime system (Amersham Biosciences, Piscataway, NJ).
An on-column procedure employing PreScission protease was used to cleave αC (233-425) from the GST tag. The released αC (233-425) was eluted with phosphate-buffered saline buffer. Complete cleavage of the GST tag was confirmed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and western blot analysis (Amersham anti-GST–horseradish peroxidase conjugate antibody). The resultant αC (233-425) was concentrated in a 5000 MWCO Vivaspin 2 Concentrator (Sartorius, Göttingen, Germany). Protein concentrations were determined using an extinction coefficient of 41480 M−1cm−1 calculated from ExPASy (www.expasy.org).
MALDI-TOF mass spectrometry kinetic assay: monitoring crosslinking reaction of glutamines in αC (233-425)
The ability of FXIIIa to crosslink each reactive glutamine in αC (233-425) to the lysine mimic GEE was monitored using our previously optimized MALDI-TOF mass spectrometry kinetic assay.47 Final concentrations of 500 nM FXIII, 17 mM GEE, 4 mM CaCl2, and MALDI assay buffer (100 mM Tris-acetate, 150 mM NaCl, and 0.1% PEG8000, pH 7.4] were incubated at 37°C in a 1.5 mL reaction tube. The FXIII was activated with bovine thrombin (8.4 U/mL final) for 10 minutes and then 200 nM D-phenylalanyl-l-prolyl-l-arginine chloromethyl ketone was added to inhibit the thrombin. The crosslinking reaction between αC (233-425) and GEE was initiated by adding a final concentration of 13.6 μM αC (233-425) to a total assay volume of 250 μL. At different time points, a 25 μL aliquot of this reaction mixture was removed and quenched in 1.6 μL of 160 mM EDTA (10 mM final). Control experiments were performed in the absence of FXIII.
All samples were subjected to proteolytic digestion with chymotrypsin and endoproteinase GluC (GluC) (Roche, Indianapolis, IN). For the digests, 6 μL of either the control or time point sample were combined with 6 μL chymotrypsin buffer (100 mM Tris-HCl, 10 mM CaCl2, pH 7.4) and 1.5 μL chymotrypsin (1 μg/μL) and incubated for 1 hour at 25°C. This digest was quenched with 2 μL 5% trifluoroacetic acid. GluC digest was performed in a similar manner with 6 μL of the sample, 6 μL GluC buffer (25 mM NH4HCO3, pH 7.8) and 1.5 μL GluC (0.05 μg/μL) incubated for 2 hours at 25°C and quenched with 5% trifluoroacetic acid. All digested samples were zip-tipped and analyzed using a MALDI-TOF mass spectrometer (Voyager-DE PRO; Applied Biosystems).47 α-Cyano-4-hydroxycinnamic acid matrix was employed for chymotrypsin digests and ferulic acid matrix for GluC. This assay was done in triplicate using αC (233-425) from 3 independent expression trials. The peak-height ratio method was used to determine the extent of crosslinking of GEE to each reactive glutamine with time. The amount of reactant left at each time was calculated as follows:
15N-HSQC NMR spectroscopy: validating αC (233-425) crosslinking with 15N-labeled substrates
FXIIIa-catalyzed crosslinking of 15N-labeled NH4Cl or 15N-labeled GEE to reactive glutamines in αC (233-425) was monitored using 15N-HSQC NMR spectroscopy.46 All assays were carried out in a total volume of 400 μL with 10% D2O added for the NMR deuterium lock. Final concentrations are presented. For crosslinking reactions with 15N-GEE, 800 nM FXIII was proteolytically activated with 21 U/mL bovine thrombin in the presence of 5 mM CaCl2 and 20 mM Borate buffer. Thrombin activity was quenched with 460 nM D-phenylalanyl-l-prolyl-l-arginine chloromethyl ketone; 10 mM 15N-GEE and 35 μM αC (233-425) were added and incubated for 1 hour at 37°C. For the 15NH4Cl exchange reactions, 400 nM FXIII was non-proteolytically activated in the presence of excess calcium (50 mM) and 20 mM Borate buffer (pH = 8) for 10 minutes at 37°C. Then, 100 mM 15NH4Cl and 40 μM αC (233-425) were added, and the mixture was incubated for 1 hour at 37°C. Ten percent D2O was added and the mixtures were transferred into Shigemi NMR tubes. All reactions were analyzed at 25°C using a single z-axis gradient triple resonance (HCN) cryoprobe run on a 700 MHz Varian Inova NMR Spectrometer. The parameters for the 1D HSQC were: number of transients = 512, number of increments = 1, number of points = 2048, and sweep width = 7022.5. For the 2D HSQC, the parameters used were: number of transients = 64, number of increments = 64, number of points = 2048, and sweep width = 7022.5. The spectra were processed using NMRPipe and nmrDraw.48
Results
Preparation and characterization of recombinant αC (233-425)
Recombinant αC (233-425) containing an N-terminal GST-tag and a PreScission protease cleavage site was expressed in E coli and purified. SDS–PAGE results and western blot analysis using an anti-GST antibody confirmed that there was no GST-αC or free GST protein detected in the αC samples. The amino acid sequence of αC (233-425) can be found in Figure 1A.
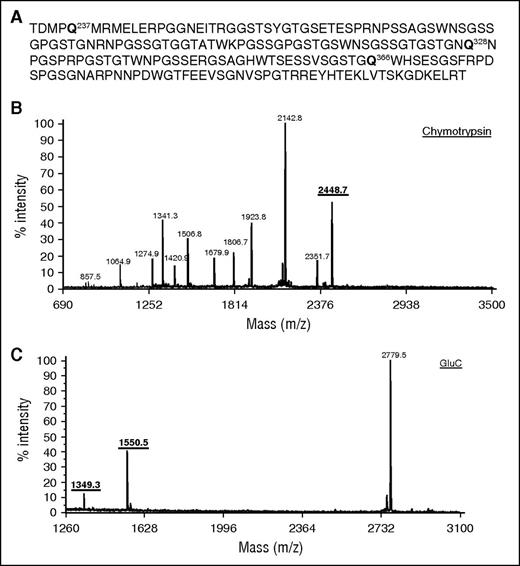
Proteolytic digests of fibrinogen αC (233-425) show three reactive glutamines in a MALDI-TOF mass spectrometry. (A) αC (233-425) sequence showing position of reactive glutamine in bold; (B) MALDI-TOF mass spectrometry spectrum of αC (233-425) digested with chymotrypsin contained a fragment with one reactive glutamine Q328 at 2448.7 m/z. (C) GluC digest of αC (233-425) showed two peaks (1550.5 m/z and 1349.3 m/z) containing Q366 and Q237, respectively, in the MALDI-TOF mass spectrometry.
Proteolytic digests of fibrinogen αC (233-425) show three reactive glutamines in a MALDI-TOF mass spectrometry. (A) αC (233-425) sequence showing position of reactive glutamine in bold; (B) MALDI-TOF mass spectrometry spectrum of αC (233-425) digested with chymotrypsin contained a fragment with one reactive glutamine Q328 at 2448.7 m/z. (C) GluC digest of αC (233-425) showed two peaks (1550.5 m/z and 1349.3 m/z) containing Q366 and Q237, respectively, in the MALDI-TOF mass spectrometry.
MALDI-TOF mass spectral analysis of separate chymotrypsin and GluC digests of αC (233-425) showed peptide fragments containing all three reactive glutamines. Theoretical protease digests (ProteinProspector; University of California San Francisco) and tandem mass spectrometry MS/MS analyses were used to identify the αC fragments. The chymotrypsin digest exhibited a peak containing Q328 (2448 m/z) (Figure 1B; and see supplemental Table 1, available on the Blood Web site), whereas the GluC digest showed peaks for Q366 (1349 m/z) and Q237 (1550 m/z) (Figure 1C; supplemental Table 1).
Monitoring crosslinking of Q237, Q328, and Q366 with GEE
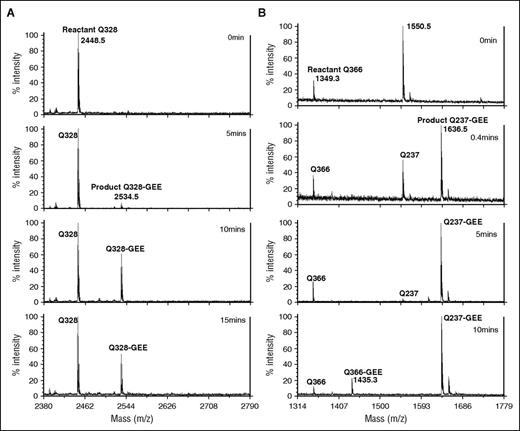
Our MALDI-TOF mass spectrometry kinetic assay was used to monitor FXIIIa-catalyzed crosslinking of each reactive αC (233-425) glutamine to the lysine mimic GEE. The crosslinking reaction was quenched at distinct time points and then digested separately with appropriate protease. A representative set of mass spectral data for a chymotrypsin (Figure 2A) and a protease GluC digest (Figure 2B) show the reactant peak containing the Q of interest and the subsequent product peak (+ 86 m/z for the new Q-GEE). No reaction products were observed in the absence of FXIIIa.
Crosslinking of reactive glutamines in fibrinogen αC (233-425) with lysine mimic GEE. Representative MALDI-TOF spectra showing crosslinking reaction between reactive glutamine in αC (233-425) and the lysine mimic GEE catalyzed by FXIIIa. (A) Chymotrypsin digest at time points 0, 5, 10, and 15 minutes show that the fragment peak containing reactive glutamine Q328 (2448.5 m/z) was crosslinked to GEE by FXIIIa to form a product peak Q328-GEE (2534.5 m/z). Each spectrum was obtained from the same starting assay and represents a quenched time point followed by a chymotrypsin digest analyzed using MALDI-TOF mass spectrometry. (B) A similar plot with GluC digest at time points 0, 0.4, 5, and 10 minutes shows reactant and product peaks containing Q237 and Q366, and their respective GEE-crosslinked products (1636.5 m/z and 1435.3 m/z). Q237 is mostly crosslinked to product by 20 seconds as compared with Q366, which was only crosslinked to product by 10 minutes.
Crosslinking of reactive glutamines in fibrinogen αC (233-425) with lysine mimic GEE. Representative MALDI-TOF spectra showing crosslinking reaction between reactive glutamine in αC (233-425) and the lysine mimic GEE catalyzed by FXIIIa. (A) Chymotrypsin digest at time points 0, 5, 10, and 15 minutes show that the fragment peak containing reactive glutamine Q328 (2448.5 m/z) was crosslinked to GEE by FXIIIa to form a product peak Q328-GEE (2534.5 m/z). Each spectrum was obtained from the same starting assay and represents a quenched time point followed by a chymotrypsin digest analyzed using MALDI-TOF mass spectrometry. (B) A similar plot with GluC digest at time points 0, 0.4, 5, and 10 minutes shows reactant and product peaks containing Q237 and Q366, and their respective GEE-crosslinked products (1636.5 m/z and 1435.3 m/z). Q237 is mostly crosslinked to product by 20 seconds as compared with Q366, which was only crosslinked to product by 10 minutes.
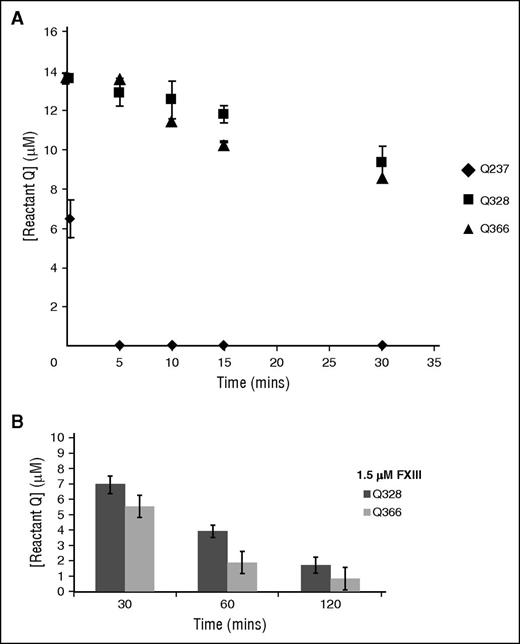
The amount of reactant left after each time point was calculated using the peak-height ratio method. As shown in Figures 2 and 3A, the peptide containing Q237 was rapidly crosslinked by 20 seconds and completely depleted by 5 minutes of reaction with GEE. FXIIIa was able to very effectively crosslink the lysine mimic GEE to the Q237 side chain amine. Q328 and Q366 could also undergo FXIII-catalyzed crosslinking with GEE but at a slower rate than Q237. A plot displaying all three αC glutamines indicates that the order of reactivity toward GEE crosslinking was Q237 ≫ Q366 ≈ Q328. Reactions with Q366 and Q328 could be brought closer to completion by increasing the FXIIIa concentration by threefold and the incubation time to 120 minutes (Figure 3B). When the original FXIIIa concentration was reduced 15-fold, the Q237 reaction rate became much slower with a 50% loss of reactant occurring by 5 minutes (data not shown). There have been previous reports on FXIIIa-catalyzed crosslinking pairs that involve α-chain glutamine residues and their respective lysine partners.23,42 Using the current assay strategy, the individual reactivities of each glutamine could, for the first time, be directly monitored.
Plot showing combined reactivities of Q237, Q328, and Q366 in fibrinogen αC (233-425). (A) A combined graph showing the rate of consumption of all three reactive glutamines Q237 (♦), Q328 (▪), and Q366 (▲). The peak-height ratio method was used to calculate the amount of reactant left in triplicate experiments and plotted as mean ± standard deviation. These reactive glutamines were ranked as Q237 ≫ Q366 ≈ Q328. (B) An increase in FXIIIa concentration by threefold shows that Q366 reacts comparably with Q328 even at higher time points.
Plot showing combined reactivities of Q237, Q328, and Q366 in fibrinogen αC (233-425). (A) A combined graph showing the rate of consumption of all three reactive glutamines Q237 (♦), Q328 (▪), and Q366 (▲). The peak-height ratio method was used to calculate the amount of reactant left in triplicate experiments and plotted as mean ± standard deviation. These reactive glutamines were ranked as Q237 ≫ Q366 ≈ Q328. (B) An increase in FXIIIa concentration by threefold shows that Q366 reacts comparably with Q328 even at higher time points.
2D 15N HSQC NMR confirms that 15N-labeled substrates crosslink to reactive glutamines
15N-labeled substrates have been used to follow reactions and conformational changes in proteins. Here, 2D HSQC NMR was used to further examine the crosslinking of 15N-labeled substrates (15N-GEE and 15NH4Cl) to the reactive glutamines on αC (233-425). With the 15N-GEE reaction, the magnetically silent 14NH2 on the αC glutamine side chains was replaced with NMR active 15NH amide product that is part of the FXIIIa-catalyzed GEE-dependent reaction (Figure 4A). The 2D HSQC spectrum of the FXIIIa-catalyzed reaction shows 3 crosspeaks with each peak corresponding to an amide 1H (8.4 to 8.1 ppm) directly attached to a 15N (114 ppm range) (Figure 4B). The observed HSQC peaks revealed that FXIIIa had successfully crosslinked 15N-GEE into the three reactive glutamines within αC (233-425). One crosspeak clearly had an intensity greater than the other two (Figure 4B). Additional HSQC studies demonstrated that 15NH4Cl could also be used as a lysine replacement (supplemental Figure 1).
Reactive glutamines in αC (233-425) are crosslinked by FXIIIa in wild-type αC and following single Q to N substitutions. (A) Reaction scheme for FXIIIa-catalyzed crosslinking reaction of 15N-GEE with reactive glutamines in αC (233-425) as monitored by NMR spectroscopy. (B) 2D 1H-15N HSQC NMR spectrum for the reactions between αC (233-425) and 15N-GEE. The 3 NMR peaks observed correspond to successful crosslinking reactions with the 3 glutamines Q237, Q328, and Q366. Each NMR peak corresponds to a proton (HN) covalently attached to an 15N-labeled nitrogen. (C) FXIIIa-catalyzed reactions between 15N-GEE and the individual αC mutants Q237N, Q328N, and Q366N as monitored by 2D 1H-15N HSQC NMR. In αC Q237N, peaks corresponding to Q328 and Q366 were seen (top). Q237 reacted fastest following either Q328N (middle) or Q366N (bottom) substitutions.
Reactive glutamines in αC (233-425) are crosslinked by FXIIIa in wild-type αC and following single Q to N substitutions. (A) Reaction scheme for FXIIIa-catalyzed crosslinking reaction of 15N-GEE with reactive glutamines in αC (233-425) as monitored by NMR spectroscopy. (B) 2D 1H-15N HSQC NMR spectrum for the reactions between αC (233-425) and 15N-GEE. The 3 NMR peaks observed correspond to successful crosslinking reactions with the 3 glutamines Q237, Q328, and Q366. Each NMR peak corresponds to a proton (HN) covalently attached to an 15N-labeled nitrogen. (C) FXIIIa-catalyzed reactions between 15N-GEE and the individual αC mutants Q237N, Q328N, and Q366N as monitored by 2D 1H-15N HSQC NMR. In αC Q237N, peaks corresponding to Q328 and Q366 were seen (top). Q237 reacted fastest following either Q328N (middle) or Q366N (bottom) substitutions.
HSQC experiments with 15N-GEE were then carried out using αC (233-425) mutants, where a single Q was replaced each time with a nonreactive N. As shown in Figure 4C, each mutant displayed 2 as opposed to the 3 crosspeaks seen in wild-type αC (233-425). A comparison of chemical shift positions and the mutant employed revealed that the strong intensity peak (at 114 ppm, 8.35 ppm) corresponds to the Q237. Q328 and Q366 occur at 113.8 ppm, 8.20 ppm and 113.9 ppm, 8.16 ppm, respectively (Figure 4B-C). The high intensity peak for Q237 matches well with the highest reactivity observed with the MALDI-TOF mass spectrometry kinetics assay, and followed by Q366 and Q328.
Discussion
Within a blood clot, FXIIIa introduces γ-glutamyl-ε-lysyl crosslinks between specific reactive glutamines and lysines located in the fibrin γ and α chains.12,13,17,41 Little is known about the individual contributions of each reactive glutamine and how FXIII selects one glutamine over another. We characterized and ranked three reactive glutamines in fibrinogen αC (233-425) for their abilities to be crosslinked by FXIIIa to the lysine mimic GEE. MALDI-TOF mass spectrometry and NMR strategies were employed that would allow us to directly measure individual reactivities.
Several studies have demonstrated that FXIIIa first introduces γ-γ crosslinks into the blood clot followed by α-α crosslinks.15,19 Putative binding sites for FXIIIa have been identified within α and γ fibrinogen chains, and such sites are located either upstream or downstream from the crosslinking glutamines.28,44 γ-γ crosslinking plays key roles in fiber appearance time and the generation of fiber density, whereas α-α crosslinking is a major determinant in fibrin clot stiffness and hinders fibrinolysis ability.16,49 Interestingly, there are far more covalent Q-K pairs involving the α chains than the γ.23 FXIIIa crosslinks fibrin γQ398 or γQ399 to γK406.19 By contrast, the αC region contains multiple reactive glutamines (Q221 [and/or Q223], Q237, Q328, and Q366) and multiple reactive lysines (K418, K448, K508, K539, K556, K563, K580, and K601).22,23,41
Curiously, there is no obvious consensus sequence for the Q-containing substrates of FXIIIa.50-52 The reactive glutamines are often found in flexible regions of the substrates. It is important to point out that not all freely available glutamines are good FXIII substrates. Fibronectin contains three glutamines toward its N-terminus but only Q3 is highly reactive, minor crosslinking involves Q7 and Q9.27 α2AP (N1) contains two reactive glutamines near the N-terminus but only Q2 is used. Reactive glutamines of α2AP and PAI-2 become crosslinked to reactive lysines (K230, K303, and K413) within αC (233-425).25 α2AP is one of the few FXIIIa substrates that can be studied employing short peptides. Sequences based on α2AP (N1, 1-15) have been used to characterize individual glutamines, evaluate roles of surrounding residues, and assess effects of peptide length.53-55 By contrast, similar sized peptides based on the fibrinogen Aα and γ chains make poor Q-substrates for FXIIIa.56,57 These observations suggest that fibrinogen chains require a larger, more protein-like environment to promote substrate specificity.
αC (233-425), containing close to 200 amino acids, has been shown to be a highly promising system for probing the FXIIIa substrate specificity toward the Aα chain.44,45 αC (233-425) has three reactive glutamines and a putative FXIIIa binding site. GEE serves successfully as a lysine mimic and targets physiological, reactive glutamines.18,55,57 Unlike spectrophotometric assays that indirectly monitor crosslinking activity or SDS–PAGE gels that provide more general conclusions about crosslinking, our mass spectrometry kinetic assay directly follows each reactive glutamine. By focusing on αC (233-425), the challenges of working with multiple competing glutamines and lysines both on fibrinogen Aα and γ chains were also avoided.
Using the MALDI-TOF mass spectrometry assay, we were able, for the first time, to rank the individual glutamine players in fibrinogen αC (233-425). Results revealed that Q237 was the most reactive, followed by Q366 and Q328. Our complementary NMR techniques validated these findings. Interestingly, the αC region is proposed to be mostly disordered and no X-ray crystallographic information is available. Despite this disordered nature, all three glutamines in αC (233-425) could still serve as FXIIIa substrates. Other FXIIIa substrates may take advantage of even more defined structural features. Fibronectin, α2AP, and PAI-2 each contain a flexible region for the reactive glutamine(s) that is further supported by a folded protein architecture. Unlike Aα-based peptides, we propose that the larger αC (233-425) exhibits some localized conformational environment that allows or promotes interactions with FXIIIa.
The αC reactivity ranking Q237 ≫ Q366 ≈ Q328 provides new information to help interpret crosslinking pairs reported recently for full length fibrinogen.23 For that study, the FXIIIa-catalyzed crosslinking reactions were followed by trypsin digests and mass spectral analysis. Wang demonstrated that Q237 could be crosslinked to more lysine pairs (αK418, αK508, αK539 αK556, and αK601) than all other reactive glutamines that were probed.23 The high reactivity we documented for Q237 suggests that this amino acid is well positioned to crosslink a variety of reactive lysines. Unlike Q237, Wang reported that αC Q366 only crosslinks to K539. It should be noted that Q328 was not identified in the Wang study. Previous investigations, however, have shown that Q328 is an important reactive glutamine.22,40 In our assay, we were able to characterize Q328 following a chymotrypsin digest. Our ability to monitor all three reactive glutamines simultaneously in αC (233-425) is enhanced by digesting each sample time point separately with chymotrypsin and GluC (Figure 1).
Using an NMR assay, we were able to confirm that FXIIIa crosslinks both 15N-GEE and 15N-NH4Cl to reactive glutamines (Figure 4; supplemental Figure 1) in a similar order as seen in the MALDI-TOF kinetic assay. Furthermore, FXIIIa still crosslinks Q237 fastest following Q328N or Q366N mutations (Figure 4). Also, in the absence of the most reactive glutamine Q237N, both Q328 and Q366 are shown to be crosslinked to GEE by FXIIIa (Figure 4). Thus, FXIIIa does not need to crosslink the fast-acting αC Q237 before moving on to the other reactive glutamines. Further mutant combinations reveal that if any of the three Qs is replaced with a nonreactive N, the other two Qs can still participate in the crosslinking reactions (Figure 4). The data collected thus far suggest there is no obvious sequentiality in FXIIIa crosslinking.
Our current results can also be considered relative to disease-associated fibrinogen truncations and mutants. In fibrinogen Otago, a major portion of the αC region (271-610) is absent and in fibrinogen Keokuk, a single point mutation changes Q328 to a stop codon generating a larger truncation.39,58 In both cases, Q221, Q223, and Q237 are still available for crosslinking by FXIIIa but Q328, Q366, and multiple reactive lysines are missing. Our mass spectrometry assay suggests that Q237 is highly reactive and the Wang studies revealed that this glutamine interacts with several lysines. FXIIIa may catalyze a heterodimeric reaction between fibrin αC Q237 and fibrin γK406. In normal fibrinogen, the α-γ heterodimer has been reported to make up 2% of the fibrin crosslink contributions, but the reactive Q and K partners have not been identified.59
In fibrinogen Seoul II, a single point mutation results in αC Q328 being replaced with proline (Q328P). With this mutation, fibrin γ-γ crosslinking occurs but α-α is impaired.40 Considering our data, Q237 and Q366 would still be available in the Seoul II patients, along with potential K residues in the more C-terminal portion of αC. αC Q237 (Q221 and/or Q223) might therefore help support the dominant fibrin γ-γ crosslinks observed in SDS–PAGE gels of fibrinogen Seoul II treated with FXIIIa and calcium.40 Multiple crosslinks involving highly reactive Q237 may, however, not be sufficient to overcome the introduction of Q328P.
Recombinant variants that model fibrinogen Seoul II revealed that Q328P/Q366P exhibited greater impairment of fibrin polymerization than the single mutants Q328P or Q366P.60 The current work with αC (233-425) suggests Q328 and Q366 exhibit similar reactivities. With each single P substitution, the pyrrole group may induce a conformational change that hinders fibrin architecture and subsequent crosslinking. A greater disruption occurs with the double mutant. Fibrin α-α crosslinking, however, was still observed and likely involved Q237, Q221, and/or Q223. Unlike working with Q328P and Q366P, our Q to N substitutions within αC (233-425) provides a more conservative strategy for understanding the contributions of the different reactive Q residues. Moreover, any glutamines still present within αC mutants can be individually monitored and kinetically ranked. Q237 always remained the most reactive glutamine but no obvious sequentiality in FXIIIa crosslinking was observed. Park et al propose that conformational problems resulting from P328 help explain the consequences of Seoul II.60 Another possibility to consider is that crosslinks containing Q328 (and/or Q366) play structural roles within the fibrin clot that differ from those of Q237, Q221, and Q223. These issues can be further probed in the future using full-length fibrinogens.
In conclusion, we have characterized and ranked three known reactive glutamines Q237, Q328, and Q366 in fibrinogen αC (233-425) using a MALDI-TOF mass spectrometry kinetics assay and a complementary 2D HSQC NMR approach. αC Q237 was shown to be the most reactive glutamine, followed by Q366 and Q328. Moreover, FXIIIa can independently crosslink these reactive glutamines. Additional studies will help better understand the individual crosslinking contributions from each reactive Q and their unique effects on clot properties. Continuing investigations on the αC region are anticipated to provide new clues about sources of FXIII substrate specificity, and their applications in future drug or assay substrate designs.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank M. Merchant and D. Wilkey for their help with MS/MS analysis; B. Lynch, B. Anokhin, and R. Billur for critical project discussions; and M. C. Yappert and N. Stolowich for help with MALDI-TOF mass spectrometry and NMR instrumentation.
This research was supported by grants from the National Institutes of Health, National Heart, Lung, and Blood Institute (R01 HL68440 and R15 HL120068), British Heart Foundation Programme Grant (RG/13/3/30104), British Heart Foundation Project Grant (PG/08/052/25172), and the Telemedicine and Advanced Technology Research Center of the US Army (W81XWH).
Authorship
Contribution: K.N.M. and M.C.M. designed the research, analyzed the data, and wrote the manuscript; K.N.M. and J.D.B. performed the research; M.C.M. supervised the research; and K.A.S., R.A.S.A., and H.P. contributed vital samples, and critically reviewed the research design and manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Muriel C. Maurer, Department of Chemistry, University of Louisville, 2320 South Brook St, Louisville, KY 40292; e-mail: muriel.maurer@louisville.edu.